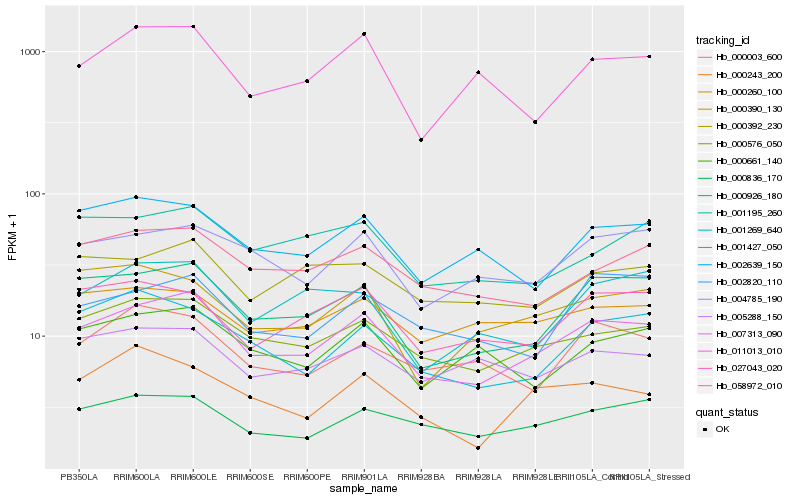
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007313_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000926_180 |
0.0770170042 |
- |
- |
proline synthetase associated protein, putative [Ricinus communis] |
| 3 |
Hb_000576_050 |
0.078616343 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 4 |
Hb_000836_170 |
0.0976347491 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 5 |
Hb_004785_190 |
0.0996006699 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 6 |
Hb_058972_010 |
0.1002130442 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 7 |
Hb_027043_020 |
0.1055109612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002639_150 |
0.1073998715 |
- |
- |
- |
| 9 |
Hb_000392_230 |
0.1093919037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001427_050 |
0.1097732552 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 11 |
Hb_011013_010 |
0.110219507 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 12 |
Hb_001195_260 |
0.1105563115 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 13 |
Hb_000661_140 |
0.1112056686 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001269_640 |
0.1119073892 |
- |
- |
hypothetical protein CISIN_1g032720mg [Citrus sinensis] |
| 15 |
Hb_000260_100 |
0.114655924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005288_150 |
0.1147979148 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_000003_600 |
0.1152880761 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1 [Jatropha curcas] |
| 18 |
Hb_002820_110 |
0.115924374 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 19 |
Hb_000243_200 |
0.1168602043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000390_130 |
0.1171182041 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B isoform X2 [Jatropha curcas] |