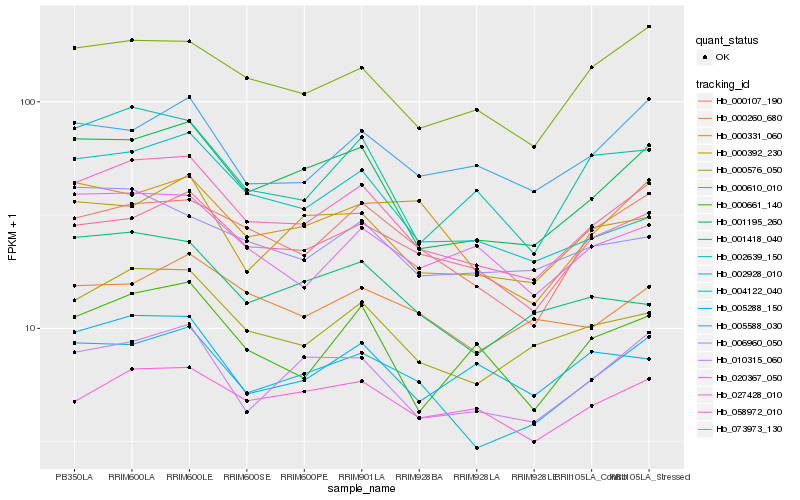
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_058972_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001195_260 |
0.0522885133 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 3 |
Hb_000576_050 |
0.0592326834 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 4 |
Hb_004122_040 |
0.0656956581 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 5 |
Hb_073973_130 |
0.0677089799 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 6 |
Hb_000107_190 |
0.0723559928 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 7 |
Hb_000610_010 |
0.0725329072 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 8 |
Hb_010315_060 |
0.0738153306 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 9 |
Hb_002639_150 |
0.0742811404 |
- |
- |
- |
| 10 |
Hb_000392_230 |
0.0756087507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002928_010 |
0.0767965823 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 12 |
Hb_020367_050 |
0.081545822 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 13 |
Hb_000661_140 |
0.083333483 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 14 |
Hb_027428_010 |
0.0837511711 |
- |
- |
alpha-1,3-mannosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_006960_050 |
0.0867532028 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 16 |
Hb_005288_150 |
0.0888335814 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_005588_030 |
0.0890473223 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 18 |
Hb_000260_680 |
0.0890613753 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 19 |
Hb_001418_040 |
0.0896215271 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 20 |
Hb_000331_060 |
0.0896388524 |
- |
- |
PREDICTED: 60S ribosomal protein L17-2-like isoform X2 [Jatropha curcas] |