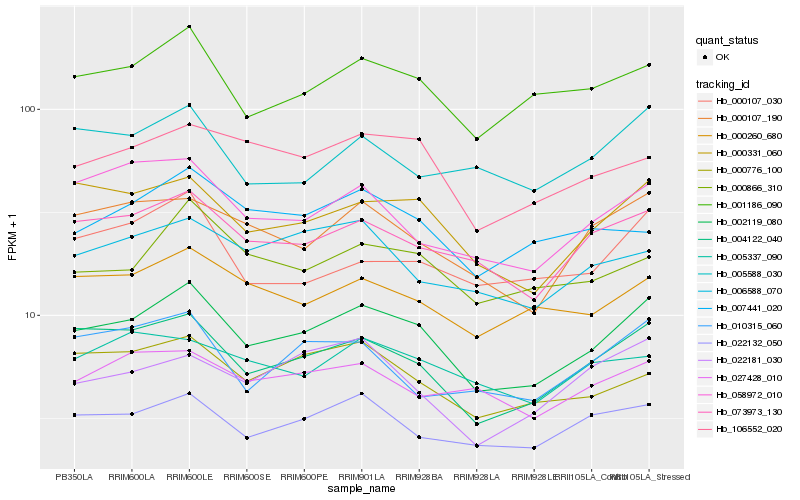
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002119_080 |
0.0 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 2 |
Hb_004122_040 |
0.0654584634 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 3 |
Hb_000331_060 |
0.0706831197 |
- |
- |
PREDICTED: 60S ribosomal protein L17-2-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_073973_130 |
0.076840115 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_001186_090 |
0.0777438394 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 6 |
Hb_000107_190 |
0.0837966702 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 7 |
Hb_022132_050 |
0.0842556238 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 8 |
Hb_106552_020 |
0.0874579087 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000260_680 |
0.0905214045 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 10 |
Hb_000866_310 |
0.0907525249 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 11 |
Hb_000776_100 |
0.0915412915 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 12 |
Hb_058972_010 |
0.0940844754 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 13 |
Hb_006588_070 |
0.0979162789 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 14 |
Hb_022181_030 |
0.1008162595 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 15 |
Hb_027428_010 |
0.1009019694 |
- |
- |
alpha-1,3-mannosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_007441_020 |
0.1009734656 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 17 |
Hb_005337_090 |
0.1022400759 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 18 |
Hb_000107_030 |
0.1026847127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010315_060 |
0.1029083504 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 20 |
Hb_005588_030 |
0.103079131 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |