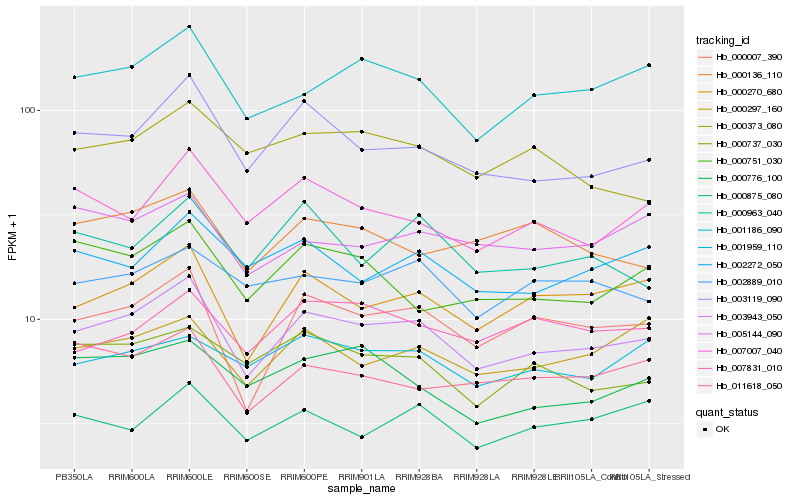
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003943_050 |
0.0 |
- |
- |
phosphoglucomutase, putative [Ricinus communis] |
| 2 |
Hb_000007_390 |
0.066644572 |
- |
- |
hypothetical protein JCGZ_18250 [Jatropha curcas] |
| 3 |
Hb_003119_090 |
0.0679429266 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 4 |
Hb_000270_680 |
0.0714733846 |
- |
- |
hypothetical protein POPTR_0015s08420g [Populus trichocarpa] |
| 5 |
Hb_001186_090 |
0.0749498884 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 6 |
Hb_007007_040 |
0.0873921683 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 7 |
Hb_000737_030 |
0.0904687187 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002272_050 |
0.0908050575 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 9 |
Hb_000297_160 |
0.0910088113 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 10 |
Hb_002889_010 |
0.0923103723 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 11 |
Hb_011618_050 |
0.0928273697 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 12 |
Hb_000776_100 |
0.0932670314 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 13 |
Hb_001959_110 |
0.09445456 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000963_040 |
0.0951922995 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 15 |
Hb_000136_110 |
0.0954800645 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 16 |
Hb_000751_030 |
0.0962359546 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 17 |
Hb_007831_010 |
0.0965515946 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 18 |
Hb_005144_090 |
0.0966193464 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 19 |
Hb_000875_080 |
0.0967771012 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 20 |
Hb_000373_080 |
0.0978473268 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |