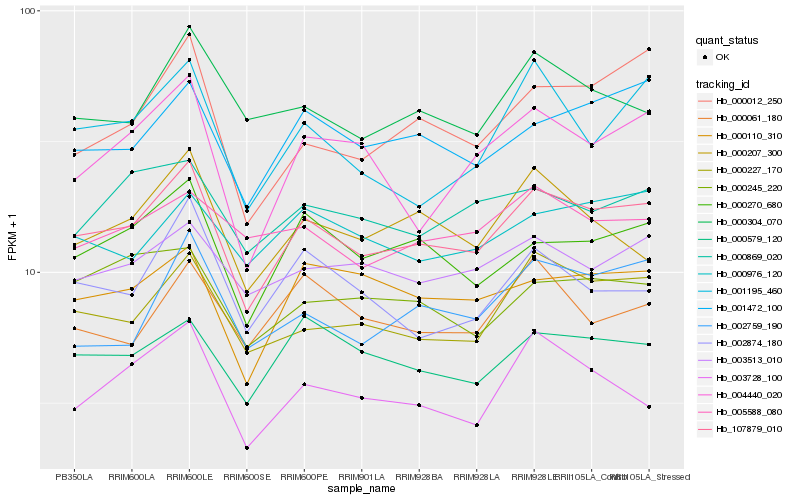
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_107879_010 |
0.0 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 2 |
Hb_000227_170 |
0.0702190319 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 3 |
Hb_000270_680 |
0.0709629877 |
- |
- |
hypothetical protein POPTR_0015s08420g [Populus trichocarpa] |
| 4 |
Hb_001472_100 |
0.0723927156 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 5 |
Hb_000579_120 |
0.077236119 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 6 |
Hb_000110_310 |
0.0782355609 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_003513_010 |
0.08128986 |
- |
- |
Structural maintenance of chromosome 1 protein, putative [Ricinus communis] |
| 8 |
Hb_000012_250 |
0.085712255 |
- |
- |
hypothetical protein L484_019524 [Morus notabilis] |
| 9 |
Hb_000304_070 |
0.0861724796 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 10 |
Hb_004440_020 |
0.0871843137 |
- |
- |
AP-3 complex subunit sigma-1, putative [Ricinus communis] |
| 11 |
Hb_000976_120 |
0.0875786227 |
- |
- |
PREDICTED: DNA polymerase zeta processivity subunit [Jatropha curcas] |
| 12 |
Hb_000869_020 |
0.0878111678 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 13 |
Hb_002759_190 |
0.088229838 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 14 |
Hb_000207_300 |
0.0883264472 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000245_220 |
0.0886703575 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 16 |
Hb_001195_460 |
0.0909222483 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_003728_100 |
0.0918966853 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002874_180 |
0.0922711349 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000061_180 |
0.0926126372 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 20 |
Hb_005588_080 |
0.0934286026 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |