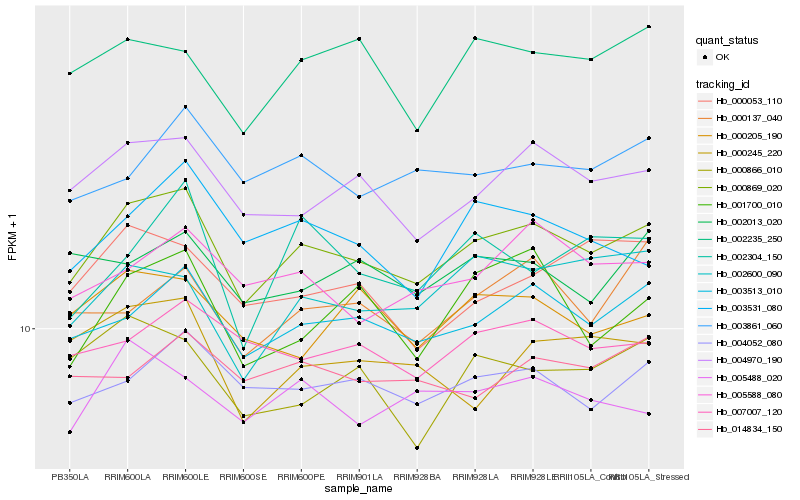
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000869_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 2 |
Hb_003513_010 |
0.055793087 |
- |
- |
Structural maintenance of chromosome 1 protein, putative [Ricinus communis] |
| 3 |
Hb_004970_190 |
0.0619010791 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_004052_080 |
0.0699307147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003861_060 |
0.0770221045 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 6 |
Hb_007007_120 |
0.0772914595 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005588_080 |
0.0773630848 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 8 |
Hb_002235_250 |
0.0775134164 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005488_020 |
0.0790353439 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 10 |
Hb_002600_090 |
0.0791323718 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_003531_080 |
0.0809109119 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000053_110 |
0.0809113736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001700_010 |
0.0810817082 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 14 |
Hb_000205_190 |
0.0812851144 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_002013_020 |
0.083192063 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 16 |
Hb_000866_010 |
0.0833099192 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000245_220 |
0.0838620823 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 18 |
Hb_014834_150 |
0.0843525888 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 1, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_000137_040 |
0.0845278612 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 20 |
Hb_002304_150 |
0.0852529848 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |