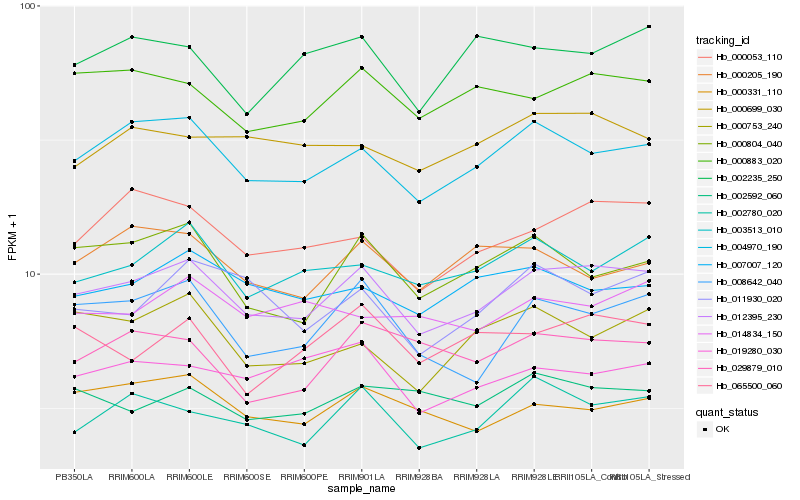
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012395_230 |
0.0 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 2 |
Hb_004970_190 |
0.0529605356 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000053_110 |
0.0670689053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008642_040 |
0.068492683 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002780_020 |
0.0693163917 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 [Jatropha curcas] |
| 6 |
Hb_000883_020 |
0.0697659541 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 7 |
Hb_003513_010 |
0.071290693 |
- |
- |
Structural maintenance of chromosome 1 protein, putative [Ricinus communis] |
| 8 |
Hb_000331_110 |
0.0749670194 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 9 |
Hb_000804_040 |
0.0753481536 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_011930_020 |
0.0768887923 |
- |
- |
PREDICTED: uncharacterized protein LOC105649787 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007007_120 |
0.0769819769 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_019280_030 |
0.0782573078 |
- |
- |
- |
| 13 |
Hb_065500_060 |
0.0783980587 |
- |
- |
PREDICTED: uncharacterized protein LOC105633723 [Jatropha curcas] |
| 14 |
Hb_000753_240 |
0.0795728686 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 15 |
Hb_000699_030 |
0.0798148137 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_002592_060 |
0.0813930551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002235_250 |
0.0820482287 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000205_190 |
0.0832726193 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_014834_150 |
0.0835355234 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 1, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_029879_010 |
0.0837942267 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |