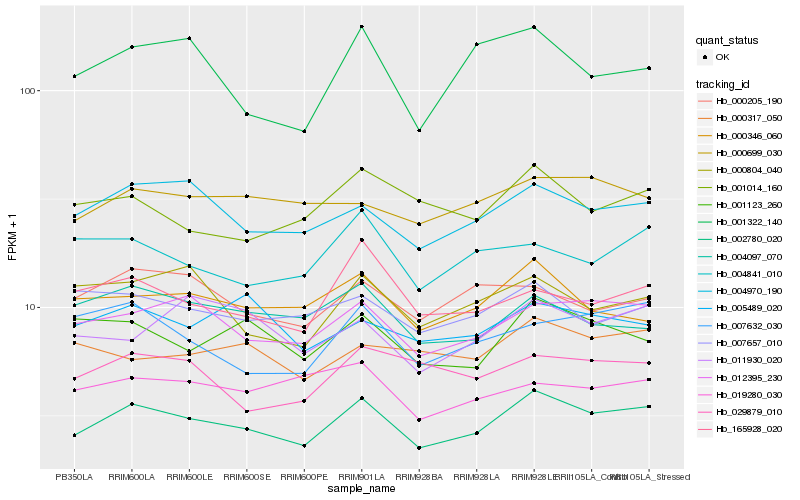
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002780_020 |
0.0 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 [Jatropha curcas] |
| 2 |
Hb_012395_230 |
0.0693163917 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 3 |
Hb_004970_190 |
0.0736347863 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000346_060 |
0.0894979333 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 5 |
Hb_001123_260 |
0.0905091215 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 6 |
Hb_000699_030 |
0.0912268964 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_007632_030 |
0.0918647805 |
- |
- |
PREDICTED: poly(A)-specific ribonuclease PARN isoform X1 [Jatropha curcas] |
| 8 |
Hb_165928_020 |
0.0926183602 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_007657_010 |
0.0927631599 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_001014_160 |
0.0928096235 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 11 |
Hb_029879_010 |
0.0940804213 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |
| 12 |
Hb_004841_010 |
0.0944269384 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000205_190 |
0.0950623032 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000804_040 |
0.0951186843 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_011930_020 |
0.0952498221 |
- |
- |
PREDICTED: uncharacterized protein LOC105649787 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004097_070 |
0.0955791535 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 17 |
Hb_000317_050 |
0.0957114361 |
- |
- |
formiminotransferase-cyclodeaminase, putative [Ricinus communis] |
| 18 |
Hb_005489_020 |
0.096880924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001322_140 |
0.0970778981 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_019280_030 |
0.0974924808 |
- |
- |
- |