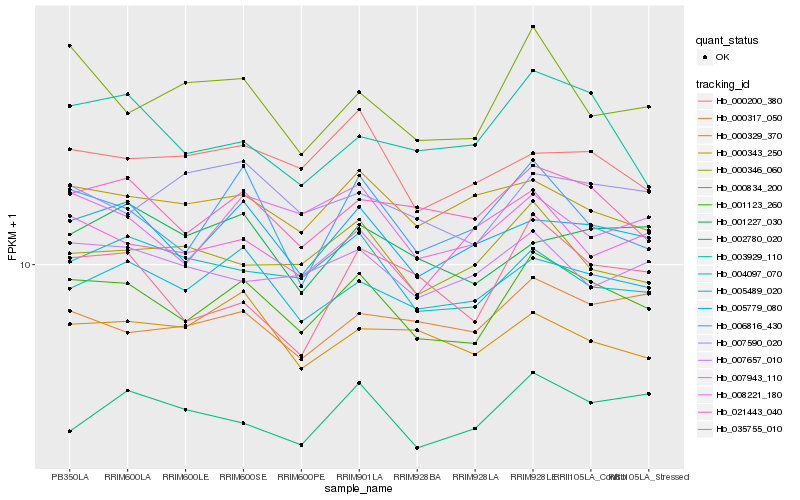
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_260 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 2 |
Hb_005779_080 |
0.0652891961 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005489_020 |
0.0684875643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_021443_040 |
0.0689458661 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 5 |
Hb_006816_430 |
0.0759120835 |
- |
- |
PREDICTED: topless-related protein 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007943_110 |
0.0776187833 |
- |
- |
PREDICTED: basic salivary proline-rich protein 3 [Jatropha curcas] |
| 7 |
Hb_004097_070 |
0.0833812125 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 8 |
Hb_000200_380 |
0.083834592 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000343_250 |
0.0839331544 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 10 |
Hb_000346_060 |
0.0849823503 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 11 |
Hb_000317_050 |
0.0853053026 |
- |
- |
formiminotransferase-cyclodeaminase, putative [Ricinus communis] |
| 12 |
Hb_007657_010 |
0.0863188762 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_003929_110 |
0.0866652846 |
- |
- |
PREDICTED: probable GTP diphosphokinase CRSH, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_008221_180 |
0.0872334743 |
- |
- |
PREDICTED: uncharacterized protein LOC105644223 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007590_020 |
0.0874464794 |
- |
- |
PREDICTED: protein DAMAGED DNA-BINDING 2 [Jatropha curcas] |
| 16 |
Hb_000834_200 |
0.0874508135 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 17 |
Hb_001227_030 |
0.0884717789 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_035755_010 |
0.0891284505 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_002780_020 |
0.0905091215 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 [Jatropha curcas] |
| 20 |
Hb_000329_370 |
0.0910179462 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |