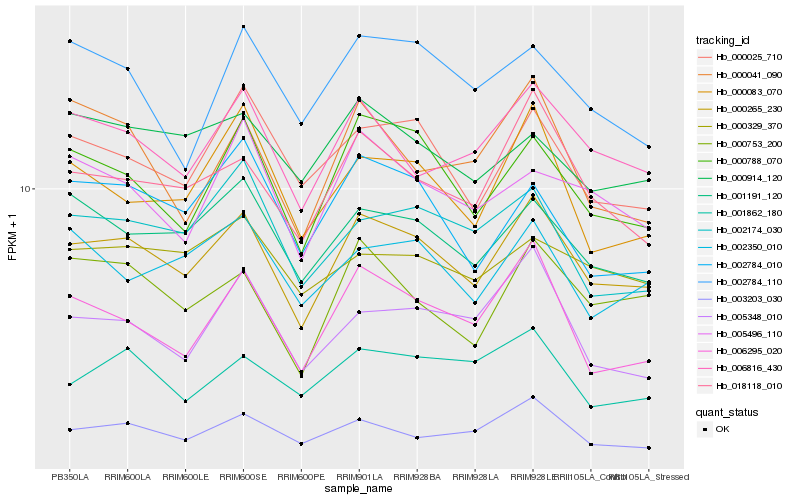
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000265_230 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105643603 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000788_070 |
0.069138093 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 3 |
Hb_018118_010 |
0.0696325334 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 4 |
Hb_006816_430 |
0.0736752952 |
- |
- |
PREDICTED: topless-related protein 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001191_120 |
0.0746162939 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000025_710 |
0.0780382698 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 7 |
Hb_003203_030 |
0.0785514812 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 8 |
Hb_000753_200 |
0.0795873424 |
- |
- |
PREDICTED: putative ATP-dependent helicase hrq1 [Jatropha curcas] |
| 9 |
Hb_006295_020 |
0.0802990437 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR isoform X1 [Jatropha curcas] |
| 10 |
Hb_000083_070 |
0.0837708104 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002350_010 |
0.0882729667 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 12 |
Hb_002174_030 |
0.0888878905 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 13 |
Hb_005496_110 |
0.0895357666 |
- |
- |
PREDICTED: uncharacterized protein LOC105107584 [Populus euphratica] |
| 14 |
Hb_001862_180 |
0.0918066436 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein ELI1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000329_370 |
0.0922751282 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 16 |
Hb_005348_010 |
0.0923520119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002784_010 |
0.0924324123 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 18 |
Hb_000041_090 |
0.0932645334 |
- |
- |
PREDICTED: uncharacterized protein LOC105641452 [Jatropha curcas] |
| 19 |
Hb_000914_120 |
0.0936915257 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002784_110 |
0.0939751074 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |