| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
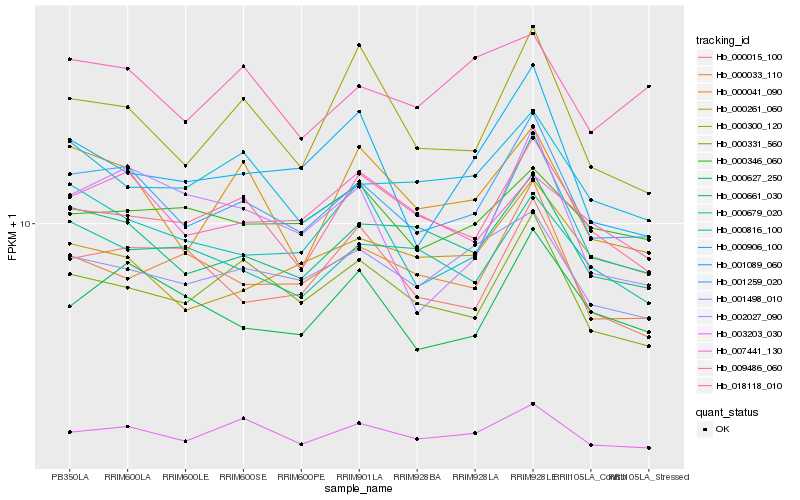
Hb_001259_020 |
0.0 |
- |
- |
PREDICTED: translocase of chloroplast 90, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000661_030 |
0.0727462981 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 3 |
Hb_003203_030 |
0.0780912158 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 4 |
Hb_000816_100 |
0.0808008433 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000331_560 |
0.0826947906 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 6 |
Hb_000627_250 |
0.0891477315 |
- |
- |
PREDICTED: uncharacterized protein LOC105632682 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001498_010 |
0.0894289546 |
- |
- |
PREDICTED: uncharacterized protein LOC105648994 [Jatropha curcas] |
| 8 |
Hb_018118_010 |
0.092309635 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 9 |
Hb_000033_110 |
0.0957835393 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 10 |
Hb_000679_020 |
0.0960938433 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 11 |
Hb_000906_100 |
0.0964281337 |
- |
- |
PREDICTED: uncharacterized protein LOC105648070 [Jatropha curcas] |
| 12 |
Hb_000041_090 |
0.0987556848 |
- |
- |
PREDICTED: uncharacterized protein LOC105641452 [Jatropha curcas] |
| 13 |
Hb_000300_120 |
0.1009304623 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000261_060 |
0.1012755131 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 15 |
Hb_009486_060 |
0.1038043063 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 16 |
Hb_000015_100 |
0.1042553815 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000346_060 |
0.1042668625 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 18 |
Hb_001089_060 |
0.1067251285 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1A isoform X1 [Jatropha curcas] |
| 19 |
Hb_002027_090 |
0.1077049647 |
- |
- |
- |
| 20 |
Hb_007441_130 |
0.1079478421 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |