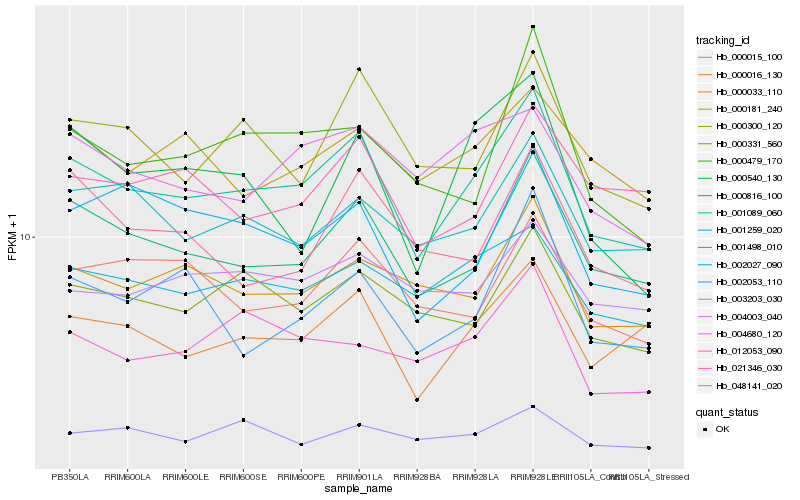
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001089_060 |
0.0 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1A isoform X1 [Jatropha curcas] |
| 2 |
Hb_000816_100 |
0.0760523321 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000016_130 |
0.0952333893 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase [Jatropha curcas] |
| 4 |
Hb_000300_120 |
0.1052154415 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000033_110 |
0.1063724247 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 6 |
Hb_001498_010 |
0.1066073743 |
- |
- |
PREDICTED: uncharacterized protein LOC105648994 [Jatropha curcas] |
| 7 |
Hb_001259_020 |
0.1067251285 |
- |
- |
PREDICTED: translocase of chloroplast 90, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000331_560 |
0.107339673 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 9 |
Hb_004680_120 |
0.1164771781 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 10 |
Hb_000015_100 |
0.1171589054 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000479_170 |
0.1187394181 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 12 |
Hb_002027_090 |
0.1193481541 |
- |
- |
- |
| 13 |
Hb_000181_240 |
0.1195649438 |
- |
- |
PREDICTED: uncharacterized protein LOC105637672 [Jatropha curcas] |
| 14 |
Hb_021346_030 |
0.1196990975 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 15 |
Hb_012053_090 |
0.1204788561 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 16 |
Hb_002053_110 |
0.1225634589 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_004003_040 |
0.1234955629 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 18 |
Hb_000540_130 |
0.1235030923 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 19 |
Hb_048141_020 |
0.1247448987 |
- |
- |
tetratricopeptide repeat-like superfamily protein, partial [Manihot esculenta] |
| 20 |
Hb_003203_030 |
0.1263756397 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |