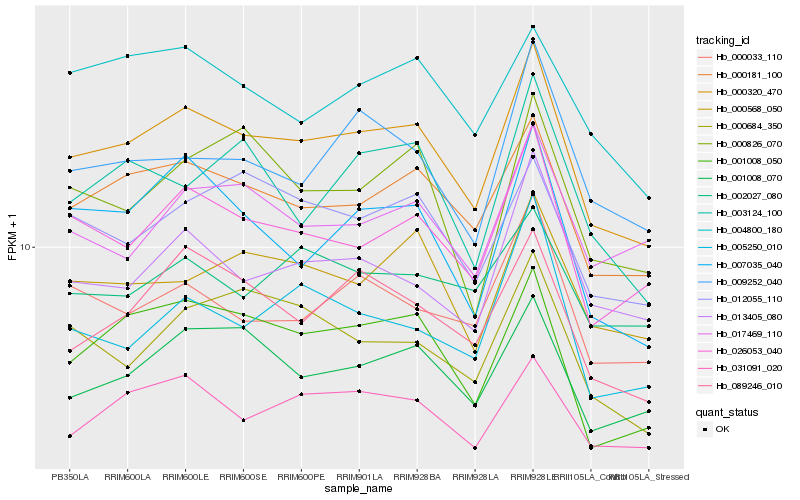
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000320_470 |
0.0 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 2 |
Hb_000181_100 |
0.0541401708 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 3 |
Hb_026053_040 |
0.0619218147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001008_050 |
0.0690903399 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 5 |
Hb_001008_070 |
0.0710543344 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 6 |
Hb_031091_020 |
0.0757106073 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 7 |
Hb_089246_010 |
0.075904175 |
- |
- |
hypothetical protein JCGZ_24395 [Jatropha curcas] |
| 8 |
Hb_000568_050 |
0.0760029149 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_013405_080 |
0.0765493435 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 10 |
Hb_007035_040 |
0.0766182492 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000033_110 |
0.0787804389 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 12 |
Hb_012055_110 |
0.0812111212 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 13 |
Hb_009252_040 |
0.0816042116 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000684_350 |
0.0853027394 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 15 |
Hb_017469_110 |
0.0871513824 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 16 |
Hb_004800_180 |
0.0875211207 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 17 |
Hb_005250_010 |
0.0876018713 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 18 |
Hb_003124_100 |
0.0883967363 |
- |
- |
- |
| 19 |
Hb_000826_070 |
0.0886180896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002027_080 |
0.0901389307 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |