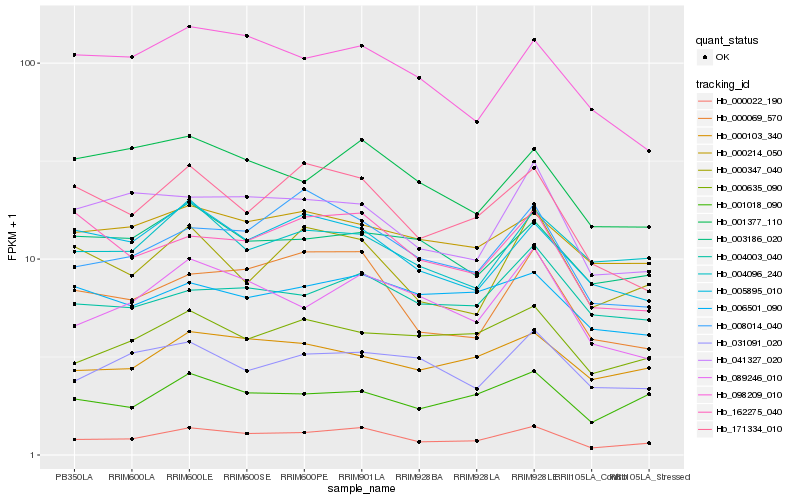
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000022_190 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22690-like [Jatropha curcas] |
| 2 |
Hb_000069_570 |
0.0892606617 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_089246_010 |
0.0937692569 |
- |
- |
hypothetical protein JCGZ_24395 [Jatropha curcas] |
| 4 |
Hb_001018_090 |
0.0957063295 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 5 |
Hb_008014_040 |
0.0967774599 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 6 |
Hb_000103_340 |
0.0989984488 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 7 |
Hb_000635_090 |
0.099220079 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 8 |
Hb_041327_020 |
0.0993929066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_171334_010 |
0.100077517 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 10 |
Hb_005895_010 |
0.1008806596 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000214_050 |
0.1010975588 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003186_020 |
0.1031037818 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001377_110 |
0.1049806906 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 14 |
Hb_004003_040 |
0.1050690874 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_031091_020 |
0.1055847955 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 16 |
Hb_006501_090 |
0.1070184844 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 17 |
Hb_004096_240 |
0.1072440602 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 18 |
Hb_162275_040 |
0.107731023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000347_040 |
0.1080604393 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 20 |
Hb_098209_010 |
0.1085580925 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |