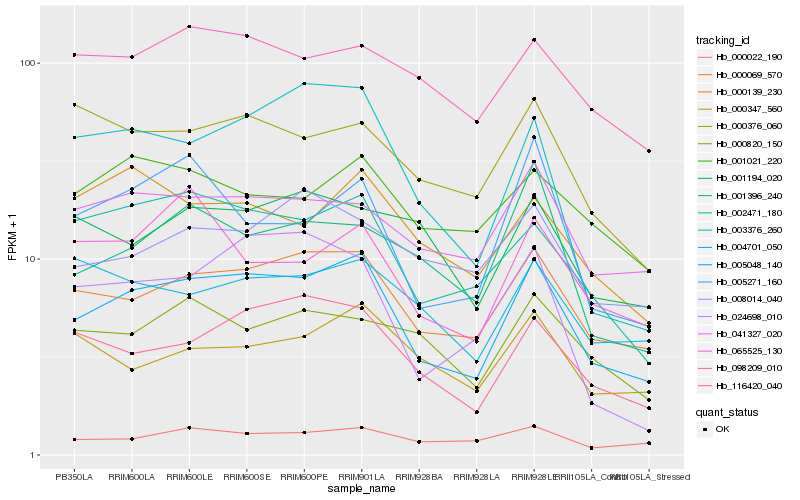
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005048_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000069_570 |
0.0896205807 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_116420_040 |
0.1170905811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002471_180 |
0.1175759568 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_041327_020 |
0.1285774798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000022_190 |
0.1350802682 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22690-like [Jatropha curcas] |
| 7 |
Hb_003376_260 |
0.135800258 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 8 |
Hb_098209_010 |
0.14059502 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 9 |
Hb_000376_060 |
0.1419056812 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005271_160 |
0.1441839433 |
- |
- |
PREDICTED: uncharacterized protein LOC105645844 [Jatropha curcas] |
| 11 |
Hb_000347_560 |
0.1447312815 |
- |
- |
- |
| 12 |
Hb_000820_150 |
0.1452760483 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 13 |
Hb_008014_040 |
0.146112271 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 14 |
Hb_004701_050 |
0.1462426234 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 15 |
Hb_001021_220 |
0.1462771338 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000139_230 |
0.1464631309 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 17 |
Hb_001396_240 |
0.1468067967 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001194_020 |
0.1477904592 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 19 |
Hb_065525_130 |
0.1488514017 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_024698_010 |
0.1496169816 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46 [Jatropha curcas] |