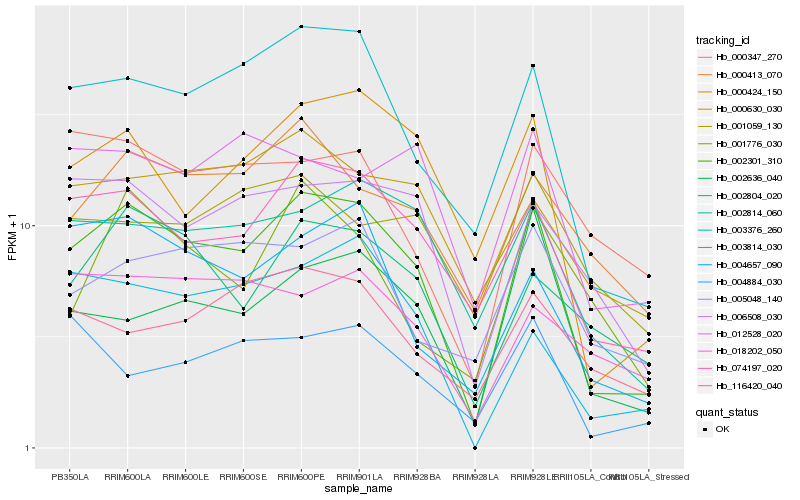
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_310 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.4 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003376_260 |
0.1205603407 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 3 |
Hb_074197_020 |
0.1270662423 |
- |
- |
PREDICTED: uncharacterized protein LOC105645948 [Jatropha curcas] |
| 4 |
Hb_000424_150 |
0.146967611 |
- |
- |
PREDICTED: copper transporter 5.1-like [Jatropha curcas] |
| 5 |
Hb_002814_060 |
0.1518278476 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 6 |
Hb_006508_030 |
0.1538825603 |
- |
- |
PREDICTED: CDPK-related kinase 1-like [Jatropha curcas] |
| 7 |
Hb_004657_090 |
0.1594724067 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 8 |
Hb_002804_020 |
0.1605267993 |
- |
- |
epsin N-terminal homology domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_000413_070 |
0.1624095633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002636_040 |
0.1662484998 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003814_030 |
0.167199411 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000630_030 |
0.1692673984 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 13 |
Hb_005048_140 |
0.1735350017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_012528_020 |
0.1770290551 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Jatropha curcas] |
| 15 |
Hb_000347_270 |
0.1786287064 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 16 |
Hb_116420_040 |
0.1792692691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004884_030 |
0.1802333608 |
- |
- |
PREDICTED: ammonium transporter 3 member 3 [Vitis vinifera] |
| 18 |
Hb_018202_050 |
0.1829832145 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A [Jatropha curcas] |
| 19 |
Hb_001776_030 |
0.1876619889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001059_130 |
0.1887884518 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |