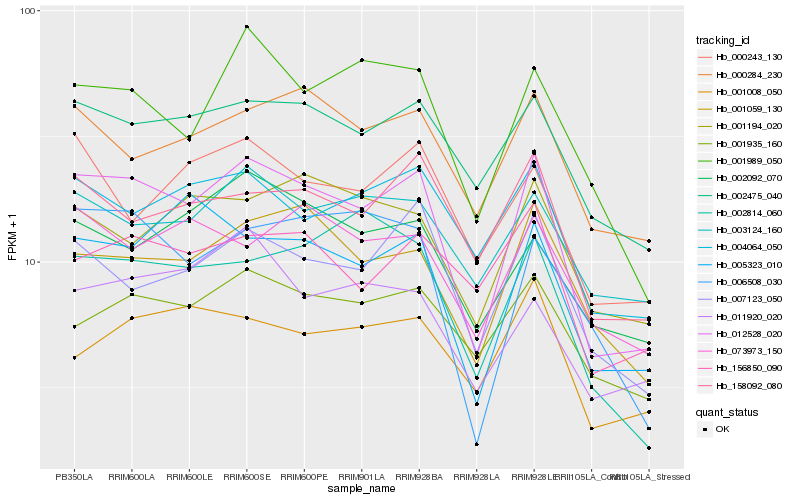
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012528_020 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Jatropha curcas] |
| 2 |
Hb_007123_050 |
0.1077270541 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_001059_130 |
0.1087118958 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002475_040 |
0.1103032106 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_005323_010 |
0.1125029452 |
- |
- |
PREDICTED: uncharacterized protein LOC105642810 [Jatropha curcas] |
| 6 |
Hb_158092_080 |
0.115917816 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 7 |
Hb_001989_050 |
0.1164194409 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 5 [Jatropha curcas] |
| 8 |
Hb_000284_230 |
0.1173726824 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 9 |
Hb_004064_050 |
0.1183679561 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 10 |
Hb_003124_160 |
0.1237430844 |
- |
- |
dynamin, putative [Ricinus communis] |
| 11 |
Hb_006508_030 |
0.1240578843 |
- |
- |
PREDICTED: CDPK-related kinase 1-like [Jatropha curcas] |
| 12 |
Hb_000243_130 |
0.1242591205 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 13 |
Hb_001935_160 |
0.1246554927 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 14 |
Hb_001194_020 |
0.1253969327 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 15 |
Hb_073973_150 |
0.1263799103 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002092_070 |
0.126442821 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 17 |
Hb_002814_060 |
0.1268644615 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 18 |
Hb_156850_090 |
0.1271556556 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 19 |
Hb_011920_020 |
0.1275439633 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 20 |
Hb_001008_050 |
0.1275510101 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |