| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
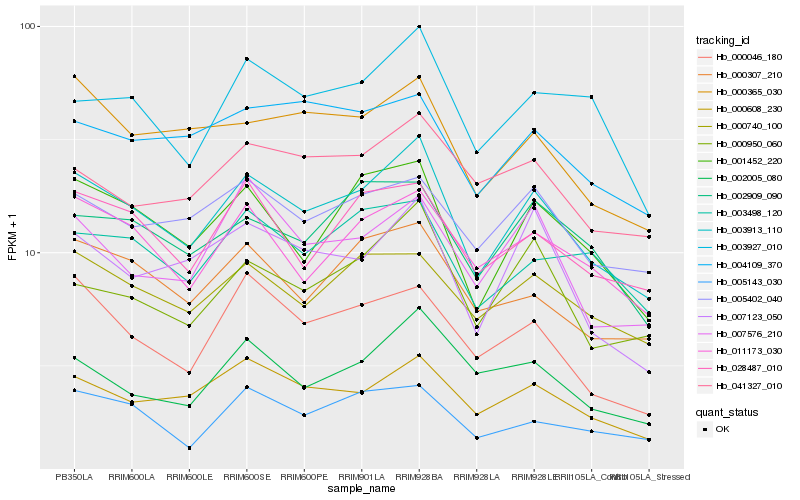
Hb_003913_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 2 |
Hb_001452_220 |
0.0791344715 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 3 |
Hb_000608_230 |
0.0819969896 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 4 |
Hb_011173_030 |
0.0882751631 |
- |
- |
pumilio, putative [Ricinus communis] |
| 5 |
Hb_000307_210 |
0.0894667726 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 6 |
Hb_000740_100 |
0.0907634823 |
- |
- |
calpain, putative [Ricinus communis] |
| 7 |
Hb_028487_010 |
0.0918005178 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000365_030 |
0.0923417441 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 9 |
Hb_000950_060 |
0.0939740157 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 10 |
Hb_005402_040 |
0.0954689893 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 11 |
Hb_007123_050 |
0.0962028134 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_003927_010 |
0.096807578 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_005143_030 |
0.0971575488 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 14 |
Hb_002909_090 |
0.097650655 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 15 |
Hb_000046_180 |
0.0986483227 |
- |
- |
PREDICTED: uncharacterized protein LOC105631840 [Jatropha curcas] |
| 16 |
Hb_003498_120 |
0.0987756714 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 17 |
Hb_002005_080 |
0.1012852623 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004109_370 |
0.102867913 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 19 |
Hb_007576_210 |
0.1038341711 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 20 |
Hb_041327_010 |
0.104958263 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |