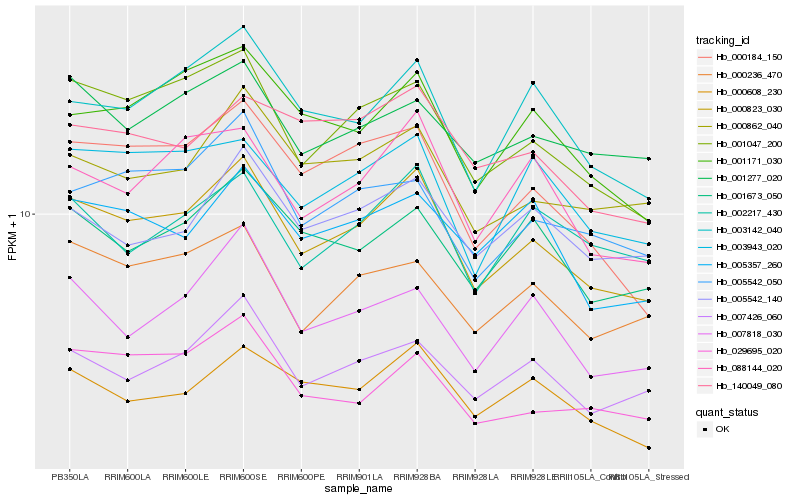
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000823_030 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 2 |
Hb_001047_200 |
0.0549841637 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 3 |
Hb_003142_040 |
0.0713459108 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 4 |
Hb_088144_020 |
0.0718053085 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_007426_060 |
0.0719525853 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000184_150 |
0.0720776131 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 7 |
Hb_001171_030 |
0.0730261854 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 8 |
Hb_000862_040 |
0.0738169532 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 9 |
Hb_001673_050 |
0.0746393864 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000608_230 |
0.0756182385 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000236_470 |
0.0758483205 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 12 |
Hb_003943_020 |
0.0761466256 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005542_050 |
0.0801533377 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 14 |
Hb_005357_260 |
0.08059064 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 15 |
Hb_005542_140 |
0.0823103398 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 16 |
Hb_002217_430 |
0.0826352577 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 17 |
Hb_007818_030 |
0.0831053971 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 18 |
Hb_001277_020 |
0.0832743572 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 19 |
Hb_029695_020 |
0.0838030984 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_140049_080 |
0.0844855906 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |