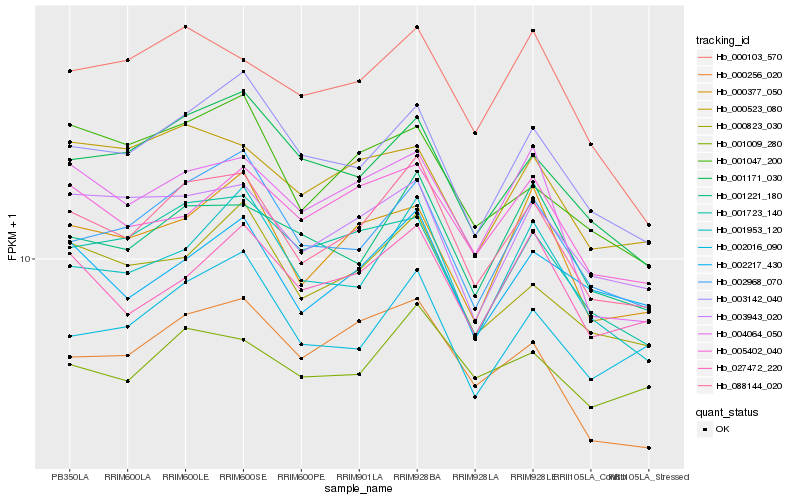
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_088144_020 |
0.0 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003142_040 |
0.0688336878 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 3 |
Hb_002968_070 |
0.0689993804 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 4 |
Hb_000823_030 |
0.0718053085 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 5 |
Hb_001953_120 |
0.0726497478 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001221_180 |
0.0743982706 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 7 |
Hb_001171_030 |
0.0746875144 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 8 |
Hb_003943_020 |
0.0772225594 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000377_050 |
0.0784897589 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 10 |
Hb_001723_140 |
0.0798458256 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 11 |
Hb_001009_280 |
0.0800476268 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 12 |
Hb_000256_020 |
0.0802193601 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 13 |
Hb_002217_430 |
0.0809522956 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 14 |
Hb_004064_050 |
0.0813256867 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 15 |
Hb_027472_220 |
0.0823310851 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000103_570 |
0.0832554424 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002016_090 |
0.0836081766 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 18 |
Hb_005402_040 |
0.0840827749 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 19 |
Hb_000523_080 |
0.0881794049 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 20 |
Hb_001047_200 |
0.088312745 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |