| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
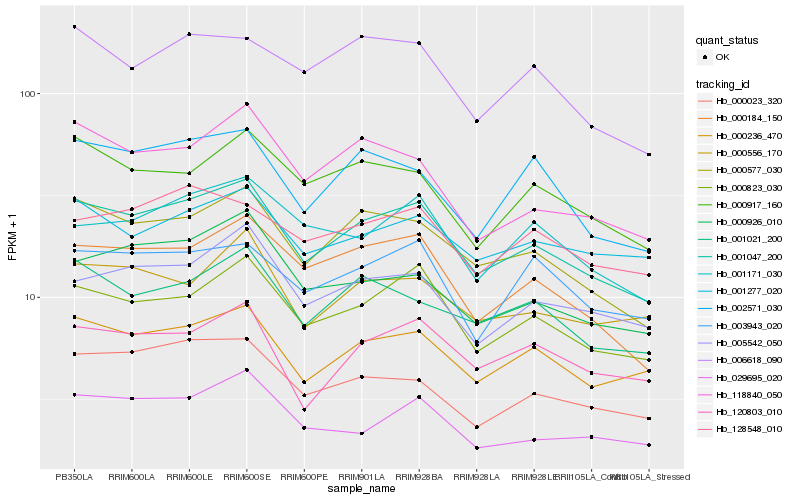
Hb_001047_200 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 2 |
Hb_000577_030 |
0.0518149659 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 3 |
Hb_000823_030 |
0.0549841637 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 4 |
Hb_000236_470 |
0.0604542765 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 5 |
Hb_000184_150 |
0.0652617673 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 6 |
Hb_001021_200 |
0.0737357682 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 7 |
Hb_005542_050 |
0.0740009504 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 8 |
Hb_002571_030 |
0.0746424864 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 9 |
Hb_000023_320 |
0.0748456599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000917_160 |
0.0778173561 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000926_010 |
0.0781712951 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 12 |
Hb_006618_090 |
0.0784184019 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 13 |
Hb_120803_010 |
0.0785887168 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein [Malus domestica] |
| 14 |
Hb_029695_020 |
0.0789109397 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003943_020 |
0.0794871369 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_118840_050 |
0.0795956655 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 17 |
Hb_000556_170 |
0.0795960019 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001277_020 |
0.0803905596 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 19 |
Hb_001171_030 |
0.0803970019 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 20 |
Hb_128548_010 |
0.0804126121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |