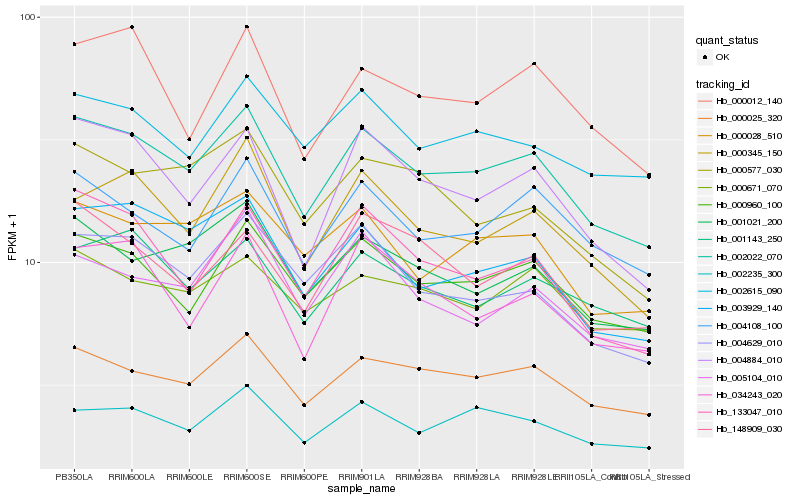
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002022_070 |
0.0 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 2 |
Hb_000960_100 |
0.0536072855 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 3 |
Hb_000025_320 |
0.0561967029 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 4 |
Hb_004884_010 |
0.0642885015 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC22 homolog [Jatropha curcas] |
| 5 |
Hb_001021_200 |
0.0658993336 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 6 |
Hb_004108_100 |
0.0675394221 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |
| 7 |
Hb_000028_510 |
0.0686914343 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 8 |
Hb_001143_250 |
0.0688162296 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 9 |
Hb_003929_140 |
0.0698313839 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_002235_300 |
0.0712636993 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002615_090 |
0.0713705136 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000345_150 |
0.0719848633 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004629_010 |
0.0733420479 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 14 |
Hb_133047_010 |
0.0734906766 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000671_070 |
0.0750287913 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 16 |
Hb_000012_140 |
0.0761088514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_148909_030 |
0.0780946575 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 18 |
Hb_034243_020 |
0.0782446161 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000577_030 |
0.0793086032 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 20 |
Hb_005104_010 |
0.0794711439 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |