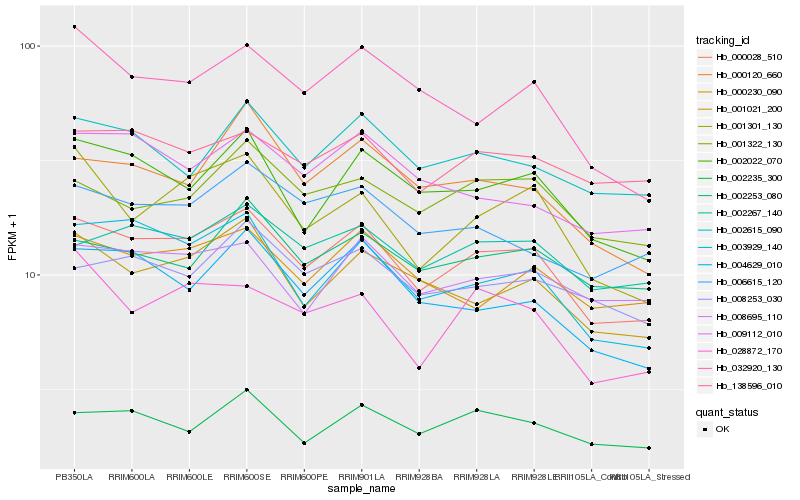
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_510 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 2 |
Hb_002022_070 |
0.0686914343 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 3 |
Hb_003929_140 |
0.0711974137 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_001021_200 |
0.0721637769 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 5 |
Hb_002267_140 |
0.0758378871 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 6 |
Hb_004629_010 |
0.0783448452 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 7 |
Hb_002615_090 |
0.0791810288 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008695_110 |
0.0797206149 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001301_130 |
0.079839597 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 10 |
Hb_000230_090 |
0.0800934242 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002235_300 |
0.0803278889 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006615_120 |
0.0817838182 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 13 |
Hb_000120_660 |
0.082560708 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 14 |
Hb_001322_130 |
0.0834324436 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 15 |
Hb_008253_030 |
0.0835642513 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 16 |
Hb_009112_010 |
0.08440873 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 17 |
Hb_028872_170 |
0.0845616722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_032920_130 |
0.0848348663 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 19 |
Hb_138596_010 |
0.0853015045 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 20 |
Hb_002253_080 |
0.0856269426 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |