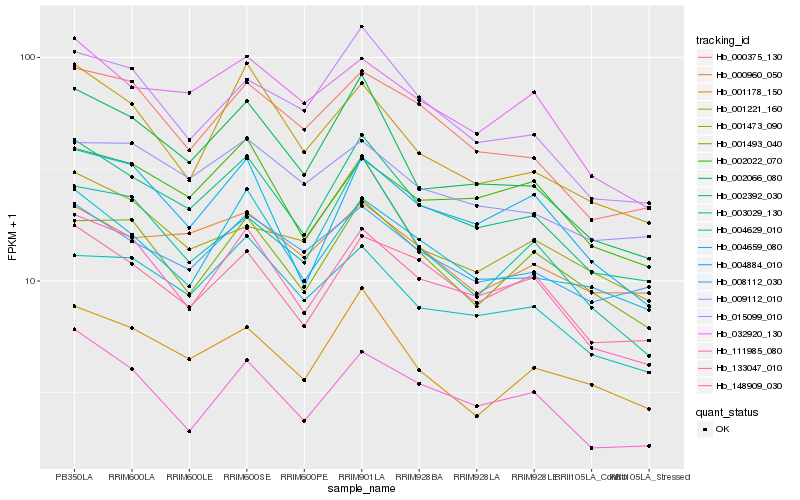
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002392_030 |
0.0 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 2 |
Hb_133047_010 |
0.0570924629 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_002066_080 |
0.0590975483 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 4 |
Hb_004629_010 |
0.0679880996 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 5 |
Hb_148909_030 |
0.0697223407 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004659_080 |
0.0701391656 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 7 |
Hb_000375_130 |
0.0711699677 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_015099_010 |
0.0712355256 |
- |
- |
PREDICTED: probable glucan 1,3-alpha-glucosidase [Jatropha curcas] |
| 9 |
Hb_032920_130 |
0.073127941 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 10 |
Hb_001178_150 |
0.0761074327 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 11 |
Hb_004884_010 |
0.0783313788 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC22 homolog [Jatropha curcas] |
| 12 |
Hb_008112_030 |
0.0790861842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001493_040 |
0.080364291 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 14 |
Hb_111985_080 |
0.0813905067 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 15 |
Hb_002022_070 |
0.0814872271 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 16 |
Hb_003029_130 |
0.0816373737 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 17 |
Hb_001473_090 |
0.081733226 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 18 |
Hb_000960_050 |
0.0820797535 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 19 |
Hb_009112_010 |
0.0822446361 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 20 |
Hb_001221_160 |
0.082538625 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |