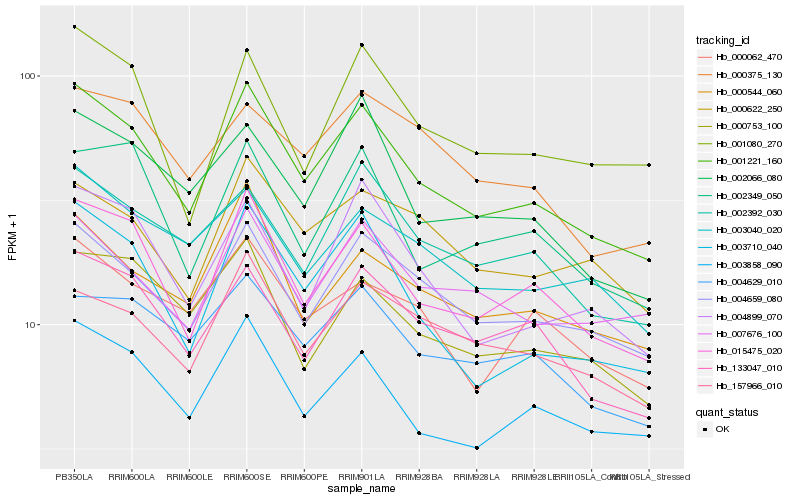
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_160 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 2 |
Hb_015475_020 |
0.0513409128 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 3 |
Hb_004659_080 |
0.0651991565 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 4 |
Hb_003858_090 |
0.0663281562 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 5 |
Hb_003710_040 |
0.070991611 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 6 |
Hb_133047_010 |
0.0756443894 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_004899_070 |
0.0795339699 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 8 |
Hb_000544_060 |
0.0821082877 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 9 |
Hb_002066_080 |
0.0824892284 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 10 |
Hb_002392_030 |
0.082538625 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 11 |
Hb_004629_010 |
0.0835721155 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 12 |
Hb_002349_050 |
0.0844045391 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 13 |
Hb_003040_020 |
0.0845678694 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 14 |
Hb_007676_100 |
0.0850625408 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 15 |
Hb_000753_100 |
0.0852749865 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001080_270 |
0.0866688816 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 17 |
Hb_157966_010 |
0.0869635035 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 18 |
Hb_000062_470 |
0.0884181543 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 19 |
Hb_000622_250 |
0.089455216 |
- |
- |
atpob1, putative [Ricinus communis] |
| 20 |
Hb_000375_130 |
0.0901069037 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |