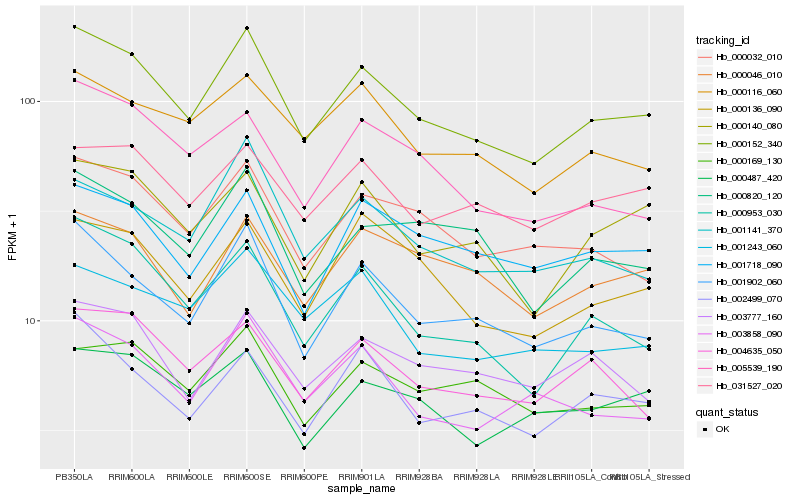
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_340 |
0.0 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 2 |
Hb_001902_060 |
0.0623598331 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000953_030 |
0.0742926018 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_000140_080 |
0.0759414598 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000032_010 |
0.0780684954 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 6 |
Hb_000116_060 |
0.0795442047 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 7 |
Hb_003858_090 |
0.0815641644 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 8 |
Hb_000136_090 |
0.0817864204 |
- |
- |
programmed cell death protein, putative [Ricinus communis] |
| 9 |
Hb_004635_050 |
0.0818880074 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 10 |
Hb_002499_070 |
0.0821175018 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001141_370 |
0.0826254289 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 12 |
Hb_031527_020 |
0.0834488027 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 13 |
Hb_003777_160 |
0.0834582237 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000046_010 |
0.0841700454 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 15 |
Hb_005539_190 |
0.0853064311 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 16 |
Hb_001718_090 |
0.0855620512 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 97 [Jatropha curcas] |
| 17 |
Hb_000820_120 |
0.0857670436 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 18 |
Hb_000169_130 |
0.0857717081 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 19 |
Hb_000487_420 |
0.0858238015 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g04390 [Jatropha curcas] |
| 20 |
Hb_001243_060 |
0.0860319864 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |