| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
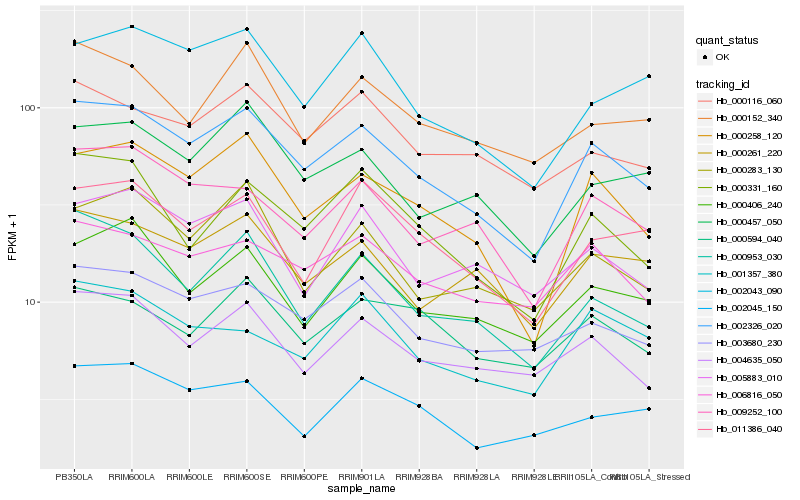
Hb_002326_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 2 |
Hb_000258_120 |
0.0627636473 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 3 |
Hb_003680_230 |
0.081115905 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 4 |
Hb_000331_160 |
0.0837147532 |
- |
- |
adenylate kinase 1 chloroplast, putative [Ricinus communis] |
| 5 |
Hb_009252_100 |
0.0848677814 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 6 |
Hb_006816_050 |
0.0853067254 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 7 |
Hb_004635_050 |
0.0859127959 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 8 |
Hb_000953_030 |
0.0862157253 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_000406_240 |
0.0898475644 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 10 |
Hb_000457_050 |
0.0915297818 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 11 |
Hb_000261_220 |
0.0916716396 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 12 |
Hb_000116_060 |
0.0930720693 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 13 |
Hb_002043_090 |
0.093437986 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 14 |
Hb_000283_130 |
0.0941548913 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 15 |
Hb_011386_040 |
0.0951406486 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005883_010 |
0.0977286124 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000152_340 |
0.097976684 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 18 |
Hb_001357_380 |
0.0984202294 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_000594_040 |
0.1010289972 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 20 |
Hb_002045_150 |
0.1017761037 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |