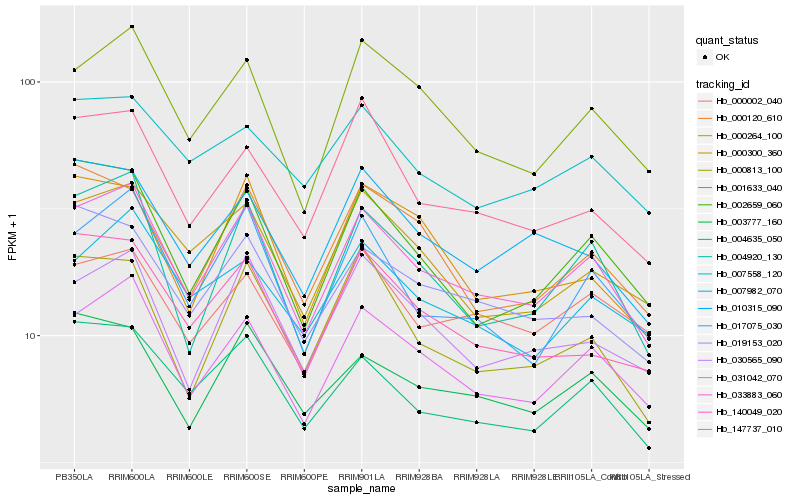
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033883_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 2 |
Hb_031042_070 |
0.0434574182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001633_040 |
0.060857559 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 4 |
Hb_004635_050 |
0.0644870615 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 5 |
Hb_017075_030 |
0.0658357682 |
- |
- |
helicase, putative [Ricinus communis] |
| 6 |
Hb_004920_130 |
0.0675751388 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_002659_060 |
0.0702800315 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 8 |
Hb_030565_090 |
0.0720345251 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 9 |
Hb_000300_360 |
0.072374933 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 10 |
Hb_003777_160 |
0.0740828305 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_147737_010 |
0.0751271782 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_019153_020 |
0.0761773214 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 13 |
Hb_140049_020 |
0.0762487764 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 14 |
Hb_010315_090 |
0.0767852674 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 15 |
Hb_007982_070 |
0.0785338435 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 16 |
Hb_000813_100 |
0.078958714 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000120_610 |
0.0796387512 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 18 |
Hb_000002_040 |
0.0810443352 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 19 |
Hb_007558_120 |
0.0831329637 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 20 |
Hb_000264_100 |
0.0843439109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |