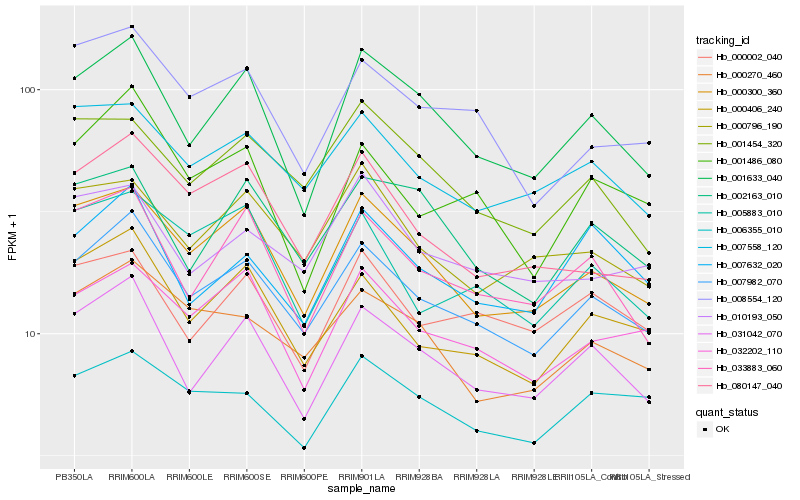
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007982_070 |
0.0 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 2 |
Hb_000300_360 |
0.0615266096 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 3 |
Hb_031042_070 |
0.0617837337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000406_240 |
0.062191511 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 5 |
Hb_000270_460 |
0.0689937292 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 6 |
Hb_007558_120 |
0.070177875 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 7 |
Hb_032202_110 |
0.0720939258 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 8 |
Hb_001633_040 |
0.0723263857 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 9 |
Hb_008554_120 |
0.0756075946 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 10 |
Hb_033883_060 |
0.0785338435 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 11 |
Hb_001486_080 |
0.0811142982 |
- |
- |
PREDICTED: eukaryotic translation initiation factor NCBP [Jatropha curcas] |
| 12 |
Hb_006355_010 |
0.0823368253 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 13 |
Hb_007632_020 |
0.083635757 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 14 |
Hb_080147_040 |
0.0838995667 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 15 |
Hb_005883_010 |
0.0843822669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000796_190 |
0.0845574051 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 17 |
Hb_000002_040 |
0.0846201879 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 18 |
Hb_001454_320 |
0.0847938469 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_010193_050 |
0.0851978141 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002163_010 |
0.0854967239 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |