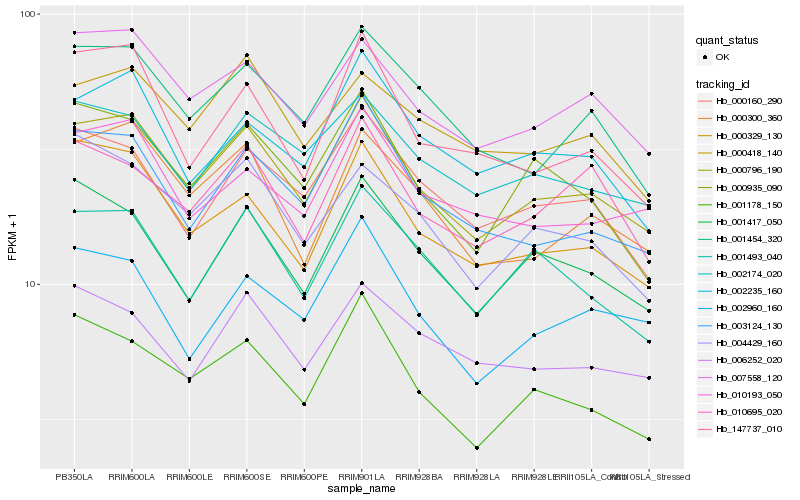
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000796_190 |
0.0 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 2 |
Hb_007558_120 |
0.0509107681 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 3 |
Hb_003124_130 |
0.0601665974 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000329_130 |
0.0608480469 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 5 |
Hb_002174_020 |
0.0613628612 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_000300_360 |
0.0629577133 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 7 |
Hb_001454_320 |
0.064076259 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001417_050 |
0.0641592017 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 9 |
Hb_001178_150 |
0.0641782371 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 10 |
Hb_001493_040 |
0.0650193906 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 11 |
Hb_000935_090 |
0.065854386 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 12 |
Hb_000160_290 |
0.0664252609 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_010193_050 |
0.0678701421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000418_140 |
0.0686070446 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 15 |
Hb_002235_160 |
0.0689618501 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 16 |
Hb_147737_010 |
0.0714488454 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_010695_020 |
0.0716402236 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 [Jatropha curcas] |
| 18 |
Hb_004429_160 |
0.0721101258 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006252_020 |
0.0727205266 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002960_160 |
0.0727236291 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |