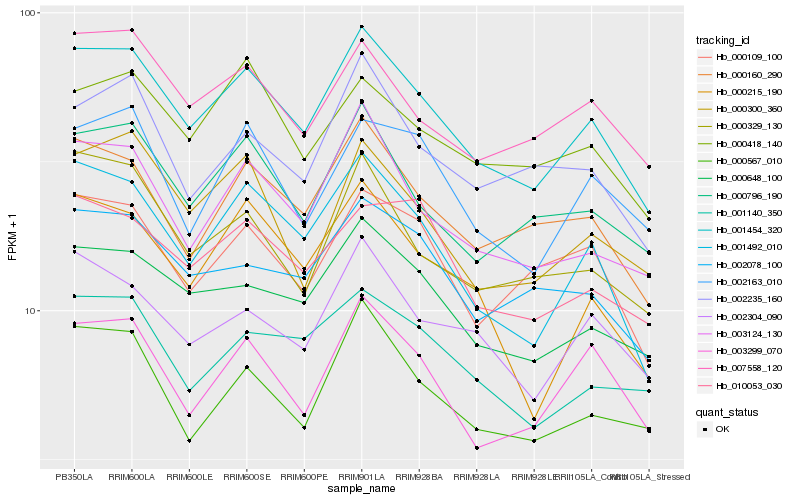
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_320 |
0.0 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001492_010 |
0.0558108235 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like isoform X2 [Populus euphratica] |
| 3 |
Hb_000648_100 |
0.0580007239 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 4 |
Hb_003124_130 |
0.0614592512 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000160_290 |
0.0632544761 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000796_190 |
0.064076259 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 7 |
Hb_007558_120 |
0.0661510379 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 8 |
Hb_002304_090 |
0.0684768771 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003299_070 |
0.0686610318 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 10 |
Hb_002078_100 |
0.0711856628 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 11 |
Hb_000300_360 |
0.0714208899 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 12 |
Hb_000567_010 |
0.0724908943 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 13 |
Hb_000109_100 |
0.0725625266 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |
| 14 |
Hb_000418_140 |
0.0727409326 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 15 |
Hb_000329_130 |
0.0733790556 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 16 |
Hb_010053_030 |
0.0747453307 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 17 |
Hb_001140_350 |
0.0751714865 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 18 |
Hb_002163_010 |
0.0752534534 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 19 |
Hb_002235_160 |
0.075658088 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 20 |
Hb_000215_190 |
0.0768192894 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |