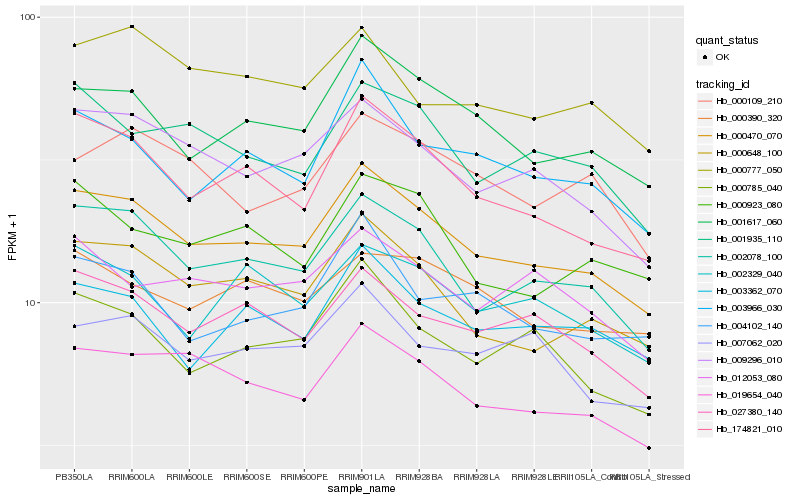
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000470_070 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 2 |
Hb_002078_100 |
0.0425734844 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 3 |
Hb_174821_010 |
0.0479252333 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 4 |
Hb_019654_040 |
0.0512319816 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000648_100 |
0.0512979622 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 6 |
Hb_009296_010 |
0.0514476126 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 7 |
Hb_001617_060 |
0.0519347573 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 8 |
Hb_027380_140 |
0.0606384453 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000785_040 |
0.0628159842 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 10 |
Hb_001935_110 |
0.0637617917 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 11 |
Hb_003966_030 |
0.0643150516 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X7 [Jatropha curcas] |
| 12 |
Hb_007062_020 |
0.0695125028 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 13 |
Hb_004102_140 |
0.0709684097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002329_040 |
0.0714961865 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 15 |
Hb_000923_080 |
0.071659633 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 16 |
Hb_000390_320 |
0.0718263814 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 17 |
Hb_012053_080 |
0.0720869421 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 18 |
Hb_003362_070 |
0.072429488 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 19 |
Hb_000109_210 |
0.0724518929 |
- |
- |
PREDICTED: threonine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000777_050 |
0.073028937 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |