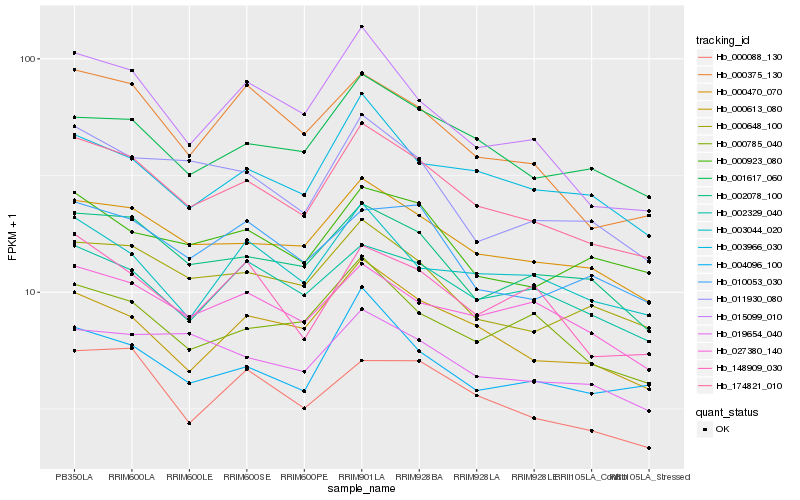
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174821_010 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 2 |
Hb_000470_070 |
0.0479252333 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 3 |
Hb_002078_100 |
0.0614129022 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 4 |
Hb_000648_100 |
0.0651079037 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 5 |
Hb_000613_080 |
0.0671929939 |
- |
- |
PREDICTED: mucin-5AC-like [Nicotiana sylvestris] |
| 6 |
Hb_001617_060 |
0.0672964495 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 7 |
Hb_003966_030 |
0.0701780869 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X7 [Jatropha curcas] |
| 8 |
Hb_000088_130 |
0.072555379 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |
| 9 |
Hb_011930_080 |
0.0726040771 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 10 |
Hb_004096_100 |
0.0728959502 |
- |
- |
PREDICTED: NADPH-dependent diflavin oxidoreductase 1 [Jatropha curcas] |
| 11 |
Hb_002329_040 |
0.0746268467 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 12 |
Hb_027380_140 |
0.0750210529 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000923_080 |
0.0750542975 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 14 |
Hb_000785_040 |
0.0751889661 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 15 |
Hb_010053_030 |
0.0757859552 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 16 |
Hb_015099_010 |
0.0762612601 |
- |
- |
PREDICTED: probable glucan 1,3-alpha-glucosidase [Jatropha curcas] |
| 17 |
Hb_019654_040 |
0.0764831688 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000375_130 |
0.0766791394 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_148909_030 |
0.0780534347 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003044_020 |
0.0797706054 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |