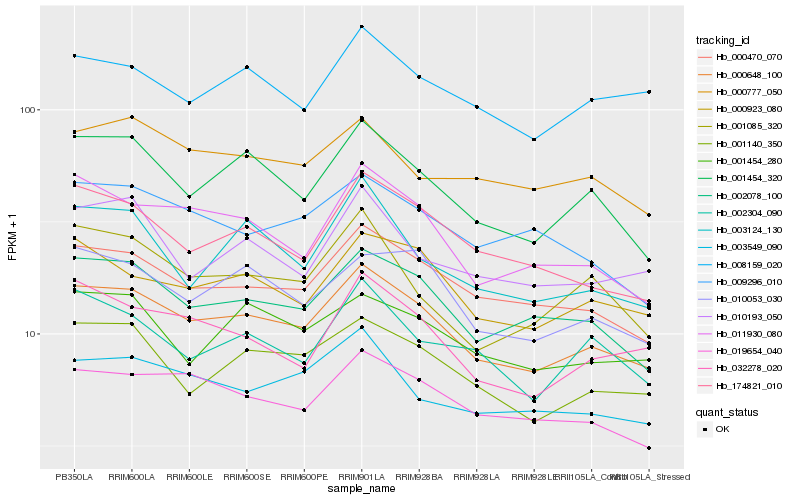
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000648_100 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 2 |
Hb_000470_070 |
0.0512979622 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 3 |
Hb_002078_100 |
0.0543203235 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 4 |
Hb_001454_320 |
0.0580007239 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_019654_040 |
0.0591607258 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000923_080 |
0.060861215 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 7 |
Hb_001140_350 |
0.0626357366 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 8 |
Hb_011930_080 |
0.0634531605 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 9 |
Hb_174821_010 |
0.0651079037 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 10 |
Hb_010053_030 |
0.065565774 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 11 |
Hb_003549_090 |
0.0693512838 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 12 |
Hb_032278_020 |
0.0713700261 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 16 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000777_050 |
0.0743244013 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 14 |
Hb_008159_020 |
0.074445035 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 15 |
Hb_001454_280 |
0.0748211388 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 16 |
Hb_002304_090 |
0.0762693243 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001085_320 |
0.0782008108 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_009296_010 |
0.0782943289 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 19 |
Hb_003124_130 |
0.0788409653 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010193_050 |
0.0789106724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |