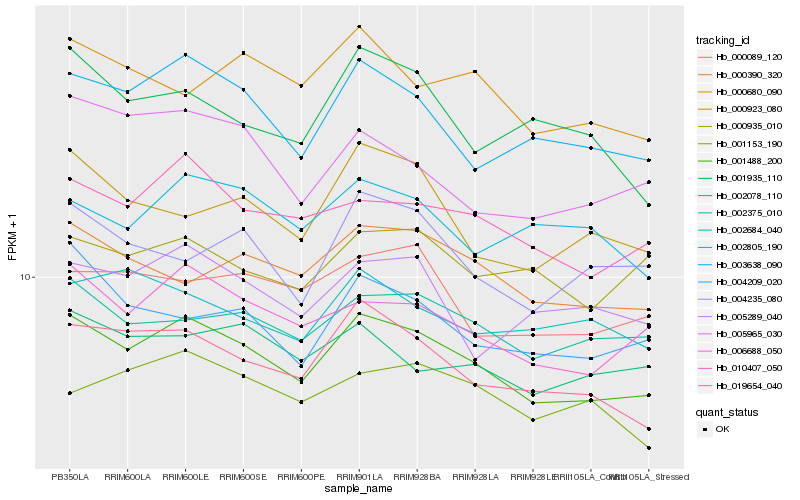
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_200 |
0.0 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_006688_050 |
0.0607447124 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX15 isoform X1 [Jatropha curcas] |
| 3 |
Hb_019654_040 |
0.0609885829 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003638_090 |
0.0619661131 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 5 |
Hb_010407_050 |
0.0633214955 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 6 |
Hb_002375_010 |
0.0707681336 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000390_320 |
0.0762253419 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 8 |
Hb_000089_120 |
0.0791580978 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_004209_020 |
0.079381447 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002078_110 |
0.0796429955 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 11 |
Hb_005289_040 |
0.0807545902 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 12 |
Hb_000923_080 |
0.08176898 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 13 |
Hb_005965_030 |
0.0820240754 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 14 |
Hb_000935_010 |
0.0820891958 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 15 |
Hb_002805_190 |
0.082557024 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 16 |
Hb_000680_090 |
0.0830744734 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 17 |
Hb_004235_080 |
0.084255229 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 18 |
Hb_001935_110 |
0.0846761619 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 19 |
Hb_002684_040 |
0.0847867824 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001153_190 |
0.0851909661 |
- |
- |
PREDICTED: DNA-directed RNA polymerase IV subunit 1 [Jatropha curcas] |