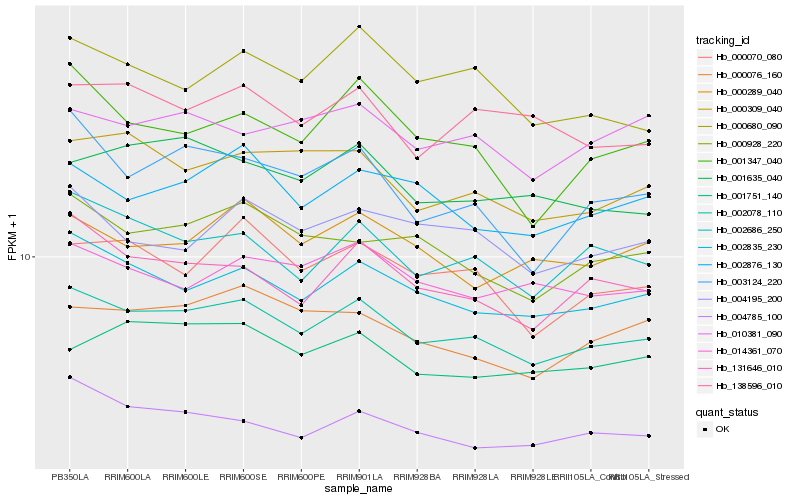
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_110 |
0.0 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 2 |
Hb_002835_230 |
0.0453647995 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_002686_250 |
0.0537981453 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 4 |
Hb_131646_010 |
0.0559097162 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000309_040 |
0.0587348875 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 6 |
Hb_000928_220 |
0.0594836871 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 7 |
Hb_000070_080 |
0.0602304605 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002876_130 |
0.0605567868 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 9 |
Hb_004785_100 |
0.0606694894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000076_160 |
0.0634720369 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 11 |
Hb_014361_070 |
0.0637789068 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 12 |
Hb_004195_200 |
0.0639734325 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 13 |
Hb_138596_010 |
0.0639793061 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 14 |
Hb_000680_090 |
0.0644108407 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 15 |
Hb_001635_040 |
0.065558005 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 16 |
Hb_010381_090 |
0.0668464055 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 17 |
Hb_000289_040 |
0.0671548378 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 18 |
Hb_001347_040 |
0.0673244898 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 19 |
Hb_001751_140 |
0.0676775516 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003124_220 |
0.0676971524 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |