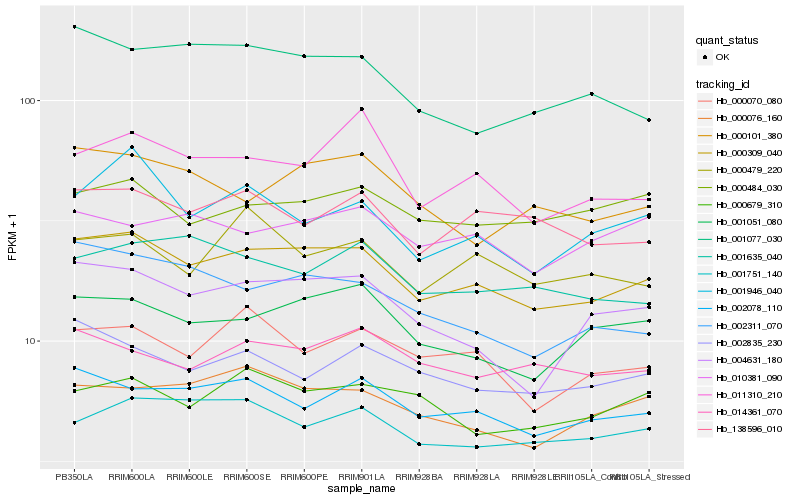
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000309_040 |
0.0 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 2 |
Hb_002078_110 |
0.0587348875 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 3 |
Hb_001051_080 |
0.06301939 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 4 |
Hb_000076_160 |
0.0671443731 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 5 |
Hb_001751_140 |
0.0703568 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 6 |
Hb_138596_010 |
0.070504925 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 7 |
Hb_002835_230 |
0.0707705597 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000070_080 |
0.0716111988 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004631_180 |
0.0726470083 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 10 |
Hb_014361_070 |
0.0731322612 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 11 |
Hb_001946_040 |
0.0736948401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011310_210 |
0.0740375643 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 13 |
Hb_001635_040 |
0.074280835 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 14 |
Hb_000101_380 |
0.074770634 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 15 |
Hb_002311_070 |
0.0753082164 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 16 |
Hb_000679_310 |
0.0755965478 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000484_030 |
0.0761093214 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Jatropha curcas] |
| 18 |
Hb_010381_090 |
0.07639426 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 19 |
Hb_000479_220 |
0.0765439623 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |
| 20 |
Hb_001077_030 |
0.077220615 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |