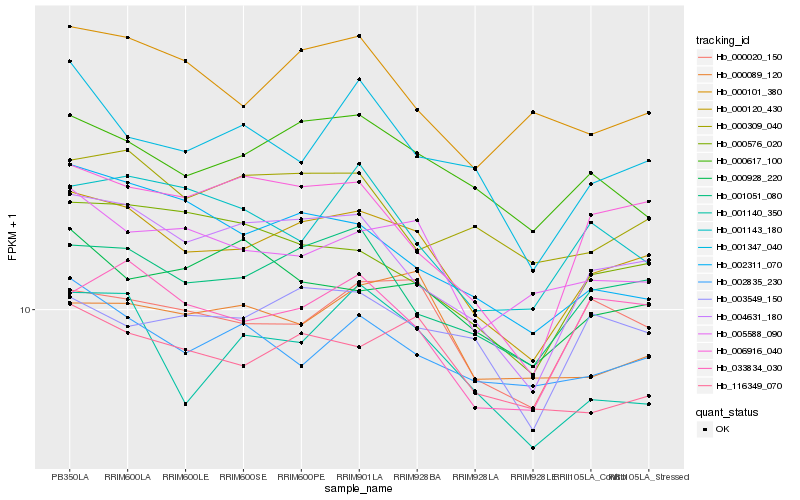
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_430 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 2 |
Hb_004631_180 |
0.0562677955 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 3 |
Hb_001051_080 |
0.058304847 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 4 |
Hb_000020_150 |
0.0670075031 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 5 |
Hb_000617_100 |
0.0676726859 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 6 |
Hb_033834_030 |
0.070521162 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_116349_070 |
0.0726776827 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 8 |
Hb_005588_090 |
0.0735310836 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 9 |
Hb_006916_040 |
0.076159027 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 10 |
Hb_003549_150 |
0.0763909491 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 11 |
Hb_000576_020 |
0.076428373 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 12 |
Hb_002835_230 |
0.078892802 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_001347_040 |
0.0798186341 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 14 |
Hb_000928_220 |
0.0805201712 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 15 |
Hb_001140_350 |
0.0816647438 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 16 |
Hb_002311_070 |
0.0817125195 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 17 |
Hb_000089_120 |
0.0821022512 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_000101_380 |
0.0826445783 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 19 |
Hb_000309_040 |
0.0831490129 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 20 |
Hb_001143_180 |
0.0832995357 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |