| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
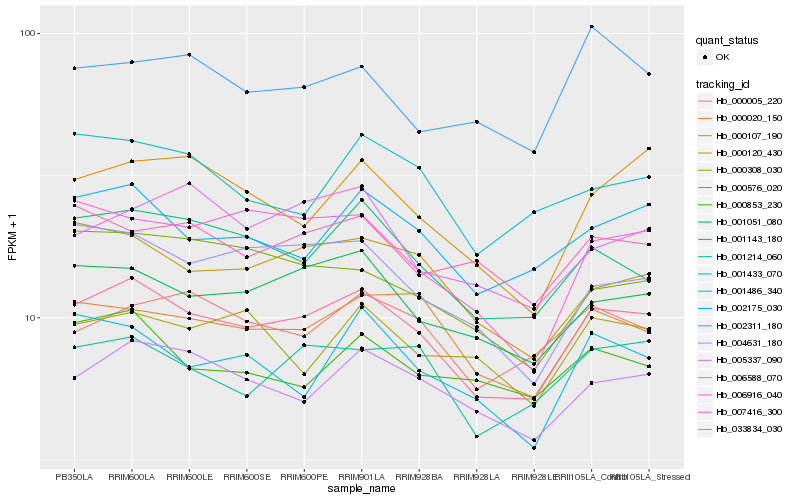
Hb_033834_030 |
0.0 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_001051_080 |
0.0616762724 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 3 |
Hb_001143_180 |
0.0652645716 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 4 |
Hb_005337_090 |
0.0655613279 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 5 |
Hb_002175_030 |
0.0658516574 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 6 |
Hb_000120_430 |
0.070521162 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 7 |
Hb_000020_150 |
0.0706730226 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 8 |
Hb_001214_060 |
0.0709972972 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC8 [Jatropha curcas] |
| 9 |
Hb_004631_180 |
0.0750357185 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 10 |
Hb_006916_040 |
0.0760002681 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 11 |
Hb_000005_220 |
0.0769788522 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 12 |
Hb_000107_190 |
0.0783471206 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 13 |
Hb_000576_020 |
0.0809709435 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 14 |
Hb_001486_340 |
0.0818774567 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 15 |
Hb_001433_070 |
0.0826795369 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 16 |
Hb_007416_300 |
0.082981316 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 17 |
Hb_000853_230 |
0.0837331688 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 18 |
Hb_006588_070 |
0.0855450749 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 19 |
Hb_000308_030 |
0.0855874138 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 20 |
Hb_002311_180 |
0.0864971232 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |