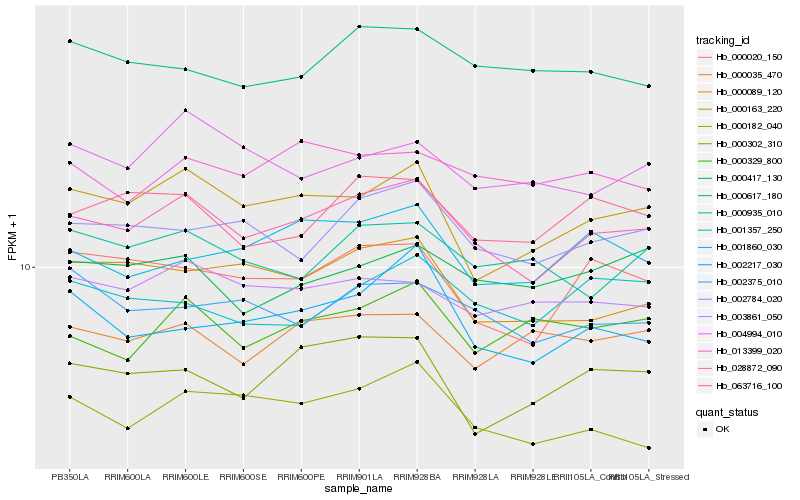
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028872_090 |
0.0 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 2 |
Hb_000163_220 |
0.0641460514 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 3 |
Hb_002784_020 |
0.0654231376 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 4 |
Hb_000020_150 |
0.0659294645 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 5 |
Hb_063716_100 |
0.0684965381 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000089_120 |
0.0734532166 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_003861_050 |
0.074276203 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 8 |
Hb_000182_040 |
0.0771586541 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 24 [Jatropha curcas] |
| 9 |
Hb_002375_010 |
0.0777076133 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 10 |
Hb_013399_020 |
0.0781663784 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 11 |
Hb_001860_030 |
0.0785906315 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 12 |
Hb_000617_180 |
0.0790590077 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 13 |
Hb_000417_130 |
0.0798280472 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 14 |
Hb_000935_010 |
0.0804055857 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 15 |
Hb_004994_010 |
0.0806436268 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 16 |
Hb_002217_030 |
0.0812040683 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000035_470 |
0.0812059806 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000302_310 |
0.081785887 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 19 |
Hb_001357_250 |
0.0827398824 |
- |
- |
PREDICTED: UPF0505 protein C16orf62 homolog [Jatropha curcas] |
| 20 |
Hb_000329_800 |
0.0836965573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |