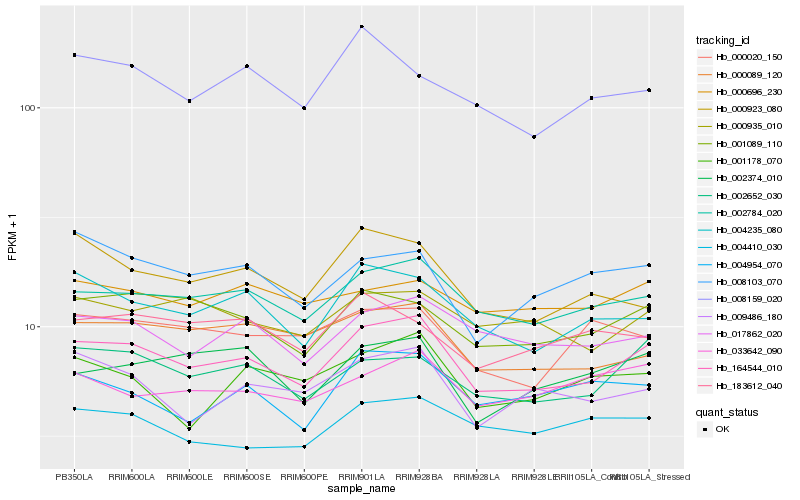
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164544_010 |
0.0 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.5 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002784_020 |
0.0556866437 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 3 |
Hb_004235_080 |
0.0716098749 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 4 |
Hb_001089_110 |
0.0719444373 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002652_030 |
0.072058297 |
- |
- |
PREDICTED: U3 small nucleolar RNA-interacting protein 2-like [Jatropha curcas] |
| 6 |
Hb_000089_120 |
0.0756847927 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_017862_020 |
0.0757261816 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 8 |
Hb_002374_010 |
0.0815466599 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_009486_180 |
0.0819375203 |
- |
- |
unknown [Glycine max] |
| 10 |
Hb_000935_010 |
0.082118068 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 11 |
Hb_008103_070 |
0.0845674812 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 12 |
Hb_033642_090 |
0.0850875742 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004954_070 |
0.0855077987 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 14 |
Hb_000923_080 |
0.0875444646 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 15 |
Hb_001178_070 |
0.0876605534 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein FBL11 [Jatropha curcas] |
| 16 |
Hb_000696_230 |
0.0876928334 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 17 |
Hb_004410_030 |
0.0884079646 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_183612_040 |
0.0889895288 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 19 |
Hb_000020_150 |
0.0890167047 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 20 |
Hb_008159_020 |
0.0896220386 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |