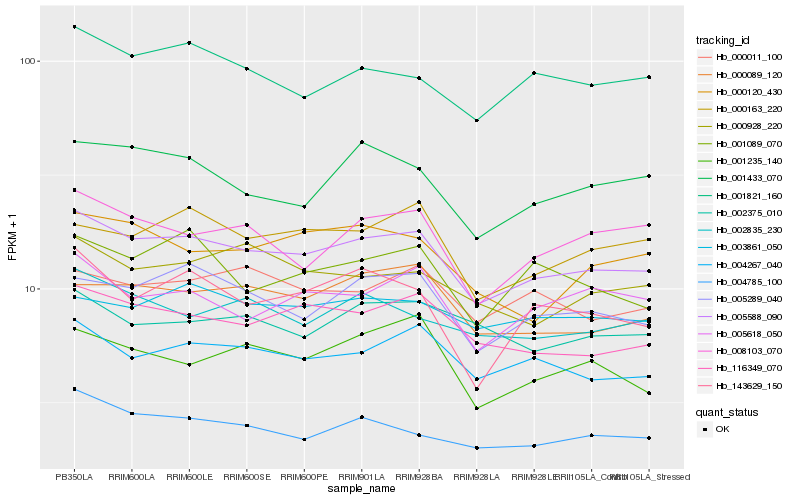
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005588_090 |
0.0 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 2 |
Hb_004267_040 |
0.0571932077 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 3 |
Hb_116349_070 |
0.0573700798 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 4 |
Hb_005289_040 |
0.0573701328 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 5 |
Hb_004785_100 |
0.0617272546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000089_120 |
0.0643930085 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_001821_160 |
0.0656537968 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 8 |
Hb_000163_220 |
0.0667787662 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 9 |
Hb_002835_230 |
0.0671309241 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_005618_050 |
0.0677491721 |
- |
- |
PREDICTED: charged multivesicular body protein 7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001433_070 |
0.067805993 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 12 |
Hb_008103_070 |
0.0694305622 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 13 |
Hb_000011_100 |
0.0701824179 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 14 |
Hb_003861_050 |
0.071344972 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 15 |
Hb_000928_220 |
0.0721209372 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 16 |
Hb_143629_150 |
0.0727776411 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |
| 17 |
Hb_001235_140 |
0.0734271019 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 18 |
Hb_000120_430 |
0.0735310836 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 19 |
Hb_001089_070 |
0.0736202423 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 20 |
Hb_002375_010 |
0.0744601696 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |