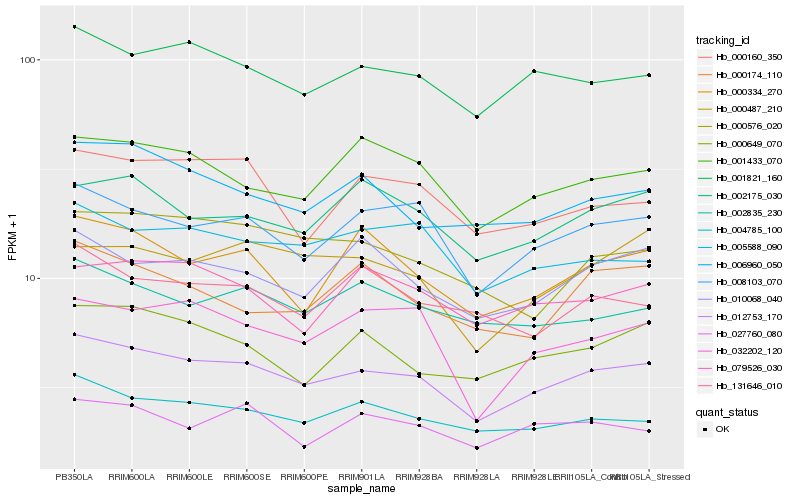
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012753_170 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_008103_070 |
0.0610395978 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 3 |
Hb_001821_160 |
0.0637866842 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 4 |
Hb_004785_100 |
0.0664273418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006960_050 |
0.0726268323 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 6 |
Hb_001433_070 |
0.0734349196 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 7 |
Hb_002175_030 |
0.0754290709 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 8 |
Hb_010068_040 |
0.0755954373 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 9 |
Hb_005588_090 |
0.076528615 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 10 |
Hb_000334_270 |
0.0767546528 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000649_070 |
0.0777780166 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 12 |
Hb_032202_120 |
0.078431839 |
- |
- |
PREDICTED: uncharacterized protein LOC105643059 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002835_230 |
0.078623149 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_079526_030 |
0.0800339276 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 15 |
Hb_000160_350 |
0.0802960624 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 16 |
Hb_000487_210 |
0.0815166306 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 17 |
Hb_000576_020 |
0.0821191347 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 18 |
Hb_027760_080 |
0.0836927335 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_131646_010 |
0.0863378688 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000174_110 |
0.0865484483 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |