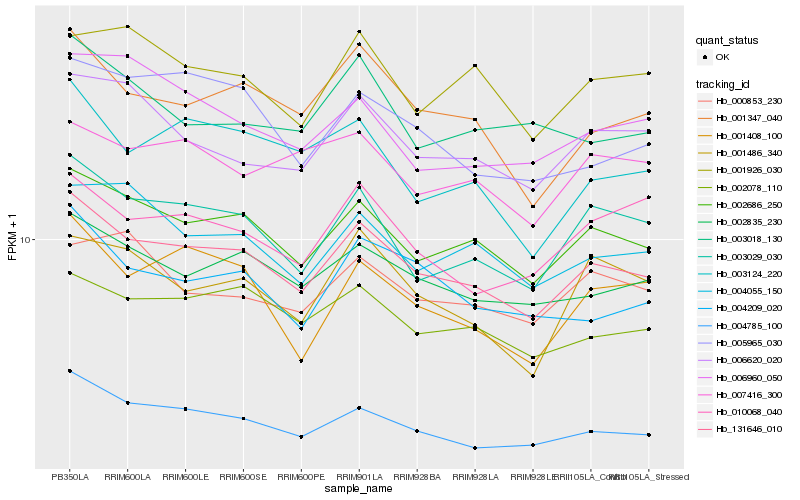
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_131646_010 |
0.0 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_002686_250 |
0.0356635007 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 3 |
Hb_004785_100 |
0.0386568894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002835_230 |
0.0494585792 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_003029_030 |
0.0543984234 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002078_110 |
0.0559097162 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 7 |
Hb_001347_040 |
0.0563120675 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 8 |
Hb_004055_150 |
0.0637407498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004209_020 |
0.0638481208 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_006620_020 |
0.0639042935 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 11 |
Hb_001408_100 |
0.0686641838 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 12 |
Hb_003124_220 |
0.0695963664 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 13 |
Hb_005965_030 |
0.0714721167 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 14 |
Hb_001486_340 |
0.0723529131 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 15 |
Hb_001926_030 |
0.0729451949 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 16 |
Hb_000853_230 |
0.0730492901 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 17 |
Hb_010068_040 |
0.0733181056 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 18 |
Hb_007416_300 |
0.0736685953 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 19 |
Hb_006960_050 |
0.0739606153 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 20 |
Hb_003018_130 |
0.0740854604 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |