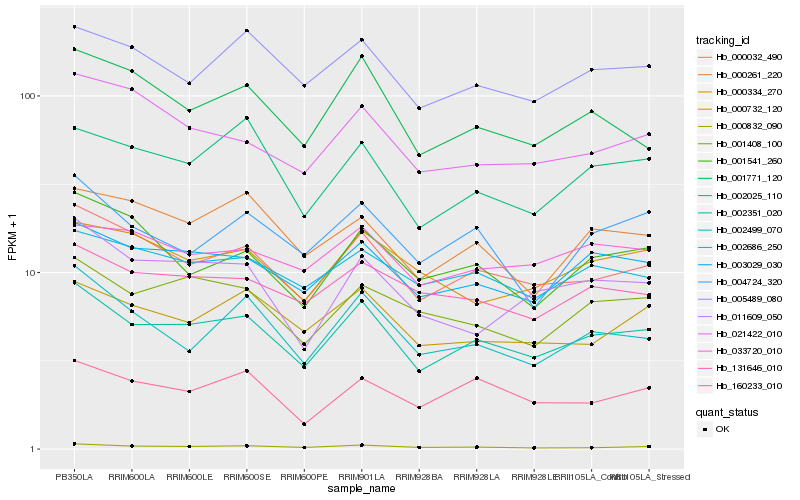
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002351_020 |
0.0 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000032_490 |
0.0582775454 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 3 |
Hb_003029_030 |
0.0668034843 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000832_090 |
0.078326106 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 5 |
Hb_005489_080 |
0.0835242767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002686_250 |
0.0864265011 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 7 |
Hb_001408_100 |
0.0869824823 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 8 |
Hb_002025_110 |
0.0871664683 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_131646_010 |
0.0877689282 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000732_120 |
0.0888875665 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001771_120 |
0.0915012351 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 12 |
Hb_002499_070 |
0.0915768924 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 13 |
Hb_160233_010 |
0.0924520845 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210 [Jatropha curcas] |
| 14 |
Hb_004724_320 |
0.0947297479 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 15 |
Hb_000261_220 |
0.0951189573 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 16 |
Hb_011609_050 |
0.0969304454 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_021422_010 |
0.097235961 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 18 |
Hb_033720_010 |
0.0972372943 |
- |
- |
PREDICTED: uncharacterized protein LOC105648843 [Jatropha curcas] |
| 19 |
Hb_000334_270 |
0.0974669543 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001541_260 |
0.0975482274 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |