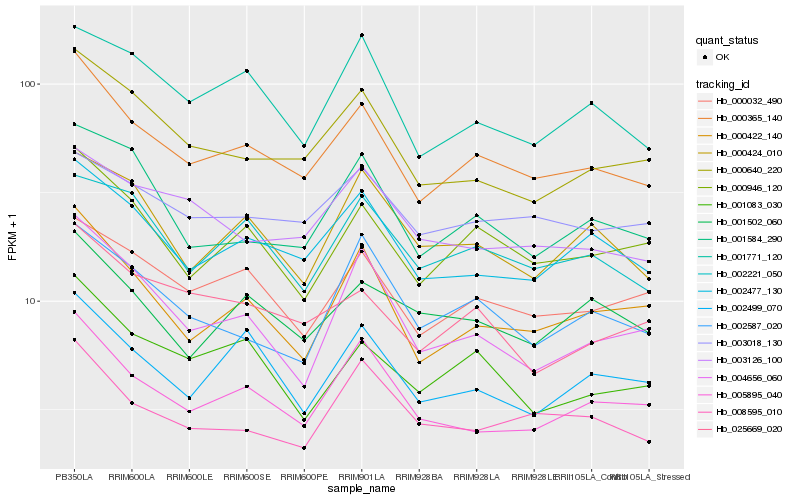
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_140 |
0.0 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 2 |
Hb_002477_130 |
0.0663485969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000946_120 |
0.0716698094 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_000032_490 |
0.0760202014 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 5 |
Hb_000422_140 |
0.0760218504 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000640_220 |
0.0765489415 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 7 |
Hb_005895_040 |
0.0837193956 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 8 |
Hb_025669_020 |
0.0866350302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001771_120 |
0.0871694513 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 10 |
Hb_001584_290 |
0.0874473554 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 11 |
Hb_002221_050 |
0.0875948497 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP65 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001083_030 |
0.0913846053 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase complex SCF subunit sconB [Jatropha curcas] |
| 13 |
Hb_008595_010 |
0.0932671445 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06710, mitochondrial [Jatropha curcas] |
| 14 |
Hb_002499_070 |
0.0942477606 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001502_060 |
0.0943744571 |
- |
- |
DNA mismatch repair protein MSH2, putative [Ricinus communis] |
| 16 |
Hb_003126_100 |
0.0945140983 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 17 |
Hb_003018_130 |
0.0946210208 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 18 |
Hb_000424_010 |
0.0948111948 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004656_060 |
0.096876237 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 20 |
Hb_002587_020 |
0.0971678774 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |