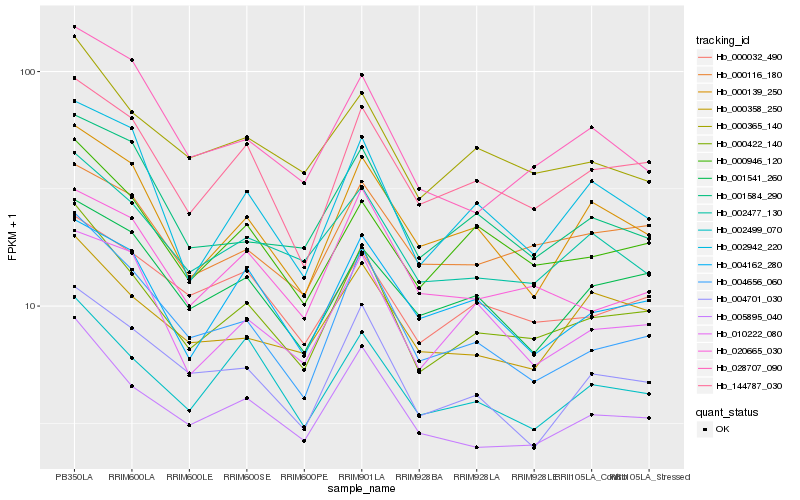
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000422_140 |
0.0 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_005895_040 |
0.0629782851 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 3 |
Hb_144787_030 |
0.0691365097 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004656_060 |
0.0694269789 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 5 |
Hb_000946_120 |
0.0726854318 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000365_140 |
0.0760218504 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 7 |
Hb_002477_130 |
0.0838042743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002499_070 |
0.0849243101 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002942_220 |
0.0868852587 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 10 |
Hb_010222_080 |
0.0870833056 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g06000 [Jatropha curcas] |
| 11 |
Hb_000139_250 |
0.0874625202 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 12 |
Hb_004701_030 |
0.0881388772 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 13 |
Hb_000032_490 |
0.0911020108 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 14 |
Hb_001584_290 |
0.0912610796 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 15 |
Hb_001541_260 |
0.0919800914 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 16 |
Hb_000116_180 |
0.0930716899 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 17 |
Hb_000358_250 |
0.0942517837 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 18 |
Hb_004162_280 |
0.0944659781 |
- |
- |
PREDICTED: uncharacterized protein LOC105628004 isoform X1 [Jatropha curcas] |
| 19 |
Hb_028707_090 |
0.0946417306 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 20 |
Hb_020665_030 |
0.0950603821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |