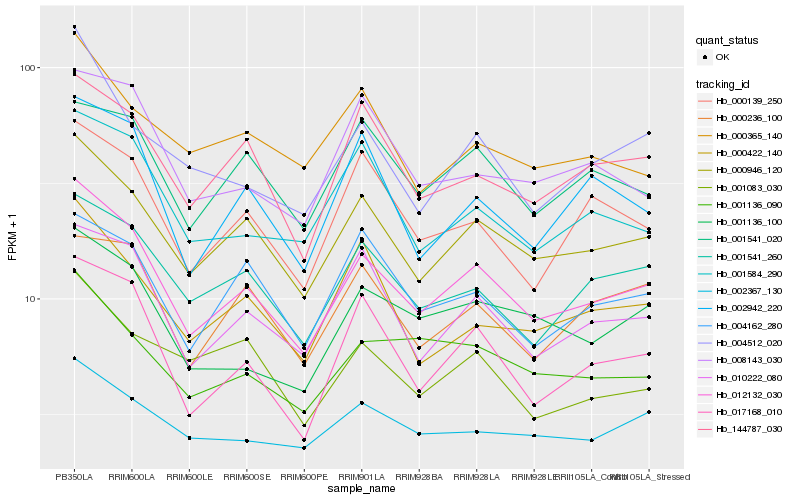
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012132_030 |
0.0 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 2 |
Hb_000946_120 |
0.0497760167 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_017168_010 |
0.077669433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010222_080 |
0.0788241601 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g06000 [Jatropha curcas] |
| 5 |
Hb_001541_260 |
0.0803905656 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 6 |
Hb_144787_030 |
0.0882613541 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001136_100 |
0.0899055016 |
- |
- |
hypothetical protein MTR_085s0013 [Medicago truncatula] |
| 8 |
Hb_000139_250 |
0.09183712 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 9 |
Hb_001584_290 |
0.0924666122 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 10 |
Hb_004512_020 |
0.0932876395 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 1 [Jatropha curcas] |
| 11 |
Hb_001136_090 |
0.093962689 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 12 |
Hb_002942_220 |
0.0943029258 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 13 |
Hb_000422_140 |
0.0959746972 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002367_130 |
0.0963754027 |
- |
- |
UNC-50 family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_001541_020 |
0.0964287272 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 16 |
Hb_000236_100 |
0.0969053048 |
- |
- |
Dynein alpha chain, flagellar outer arm [Gossypium arboreum] |
| 17 |
Hb_004162_280 |
0.0989620417 |
- |
- |
PREDICTED: uncharacterized protein LOC105628004 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000365_140 |
0.1004323488 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 19 |
Hb_001083_030 |
0.1009110917 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase complex SCF subunit sconB [Jatropha curcas] |
| 20 |
Hb_008143_030 |
0.1016436926 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X2 [Jatropha curcas] |