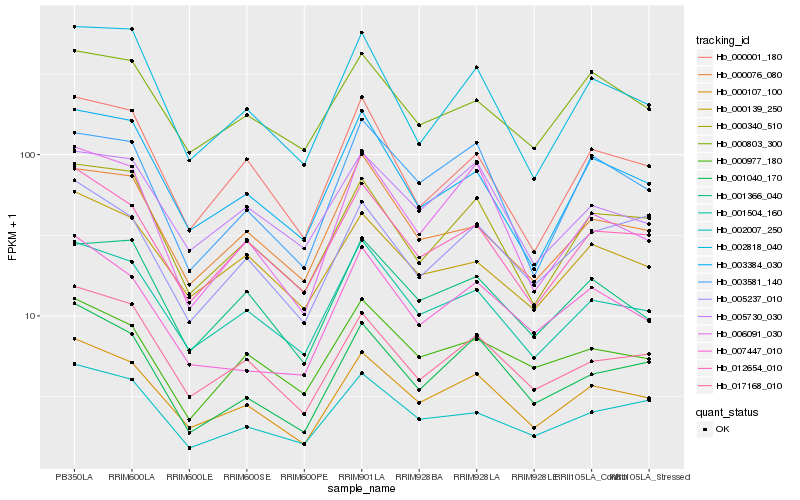
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_100 |
0.0 |
- |
- |
hypothetical protein JCGZ_16208 [Jatropha curcas] |
| 2 |
Hb_001504_160 |
0.0595839709 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 3 |
Hb_012654_010 |
0.0600034416 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 4 |
Hb_017168_010 |
0.0666412261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000340_510 |
0.0708092586 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 6 |
Hb_001040_170 |
0.0768712757 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_001366_040 |
0.0799803288 |
transcription factor |
TF Family: SNF2 |
PREDICTED: endoribonuclease Dicer homolog 1 [Jatropha curcas] |
| 8 |
Hb_000139_250 |
0.0806917409 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 9 |
Hb_002007_250 |
0.0825815066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003384_030 |
0.0832012282 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit beta-like [Jatropha curcas] |
| 11 |
Hb_005730_030 |
0.0852389364 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 12 |
Hb_000001_180 |
0.0861395836 |
- |
- |
PREDICTED: uncharacterized protein LOC105648978 [Jatropha curcas] |
| 13 |
Hb_002818_040 |
0.0864646616 |
- |
- |
tRNA synthetase-related family protein [Populus trichocarpa] |
| 14 |
Hb_005237_010 |
0.0894241598 |
- |
- |
PREDICTED: uncharacterized protein LOC105630921 [Jatropha curcas] |
| 15 |
Hb_000977_180 |
0.0894491636 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006091_030 |
0.0897952629 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog C-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000076_080 |
0.0898922216 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 18 |
Hb_007447_010 |
0.0905515893 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB3 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000803_300 |
0.0916650502 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 20 |
Hb_003581_140 |
0.0917677511 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1-like [Nelumbo nucifera] |