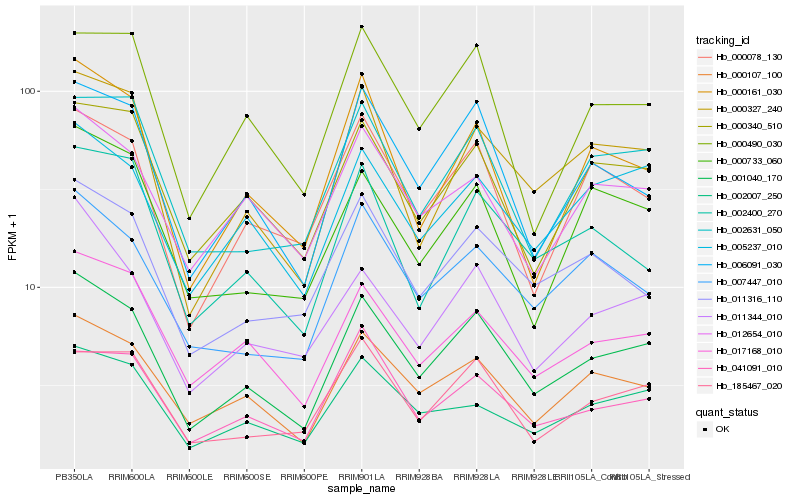
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001040_170 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_000107_100 |
0.0768712757 |
- |
- |
hypothetical protein JCGZ_16208 [Jatropha curcas] |
| 3 |
Hb_006091_030 |
0.0842448769 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog C-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_017168_010 |
0.0845782911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000327_240 |
0.0881881559 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 6 |
Hb_005237_010 |
0.0917181884 |
- |
- |
PREDICTED: uncharacterized protein LOC105630921 [Jatropha curcas] |
| 7 |
Hb_000340_510 |
0.0919091702 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 8 |
Hb_007447_010 |
0.0964705991 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB3 isoform X3 [Jatropha curcas] |
| 9 |
Hb_002631_050 |
0.09756499 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 10 |
Hb_011316_110 |
0.0979972285 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 11 |
Hb_002007_250 |
0.0989305047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000078_130 |
0.099212242 |
- |
- |
- |
| 13 |
Hb_011344_010 |
0.0995297493 |
- |
- |
PREDICTED: NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Jatropha curcas] |
| 14 |
Hb_012654_010 |
0.1001884715 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 15 |
Hb_185467_020 |
0.1002274611 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 16 |
Hb_000161_030 |
0.1008197236 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP8 [Jatropha curcas] |
| 17 |
Hb_000490_030 |
0.1021334347 |
- |
- |
DNA-directed RNA polymerase II subunit, putative [Ricinus communis] |
| 18 |
Hb_002400_270 |
0.1044185373 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_041091_010 |
0.1057530815 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000733_060 |
0.1059606705 |
- |
- |
PREDICTED: protein GUCD1 isoform X1 [Jatropha curcas] |